10.4 Drug Resistance
Learning Objectives
- Explain the concept of drug resistance
- Describe how microorganisms develop or acquire drug resistance
- Describe the different mechanisms of antimicrobial drug resistance
Antimicrobial resistance is not a new phenomenon. In nature, microbes are constantly evolving in order to overcome the antimicrobial compounds produced by other microorganisms. Human development of antimicrobial drugs and their widespread clinical use has simply provided another selective pressure that promotes further evolution. Several important factors can accelerate the evolution of drug resistance. These include the overuse and misuse of antimicrobials, inappropriate use of antimicrobials, subtherapeutic dosing, and patient noncompliance with the recommended course of treatment.
Exposure of a pathogen to an antimicrobial compound can select for chromosomal mutations conferring resistance, which can be transferred vertically to subsequent microbial generations and eventually become predominant in a microbial population that is repeatedly exposed to the antimicrobial. Alternatively, many genes responsible for drug resistance are found on plasmids or in transposons that can be transferred easily between microbes through horizontal gene tranfer. Small pieces of DNA called transposons also have the ability to move resistance genes between plasmids and chromosomes to further promote the spread of resistance.
Mechanisms for Drug Resistance
There are several common mechanisms for drug resistance, which are summarized in Figure 10.9. These mechanisms include enzymatic modification of the drug, modification of the antimicrobial target, and prevention of drug penetration or accumulation.
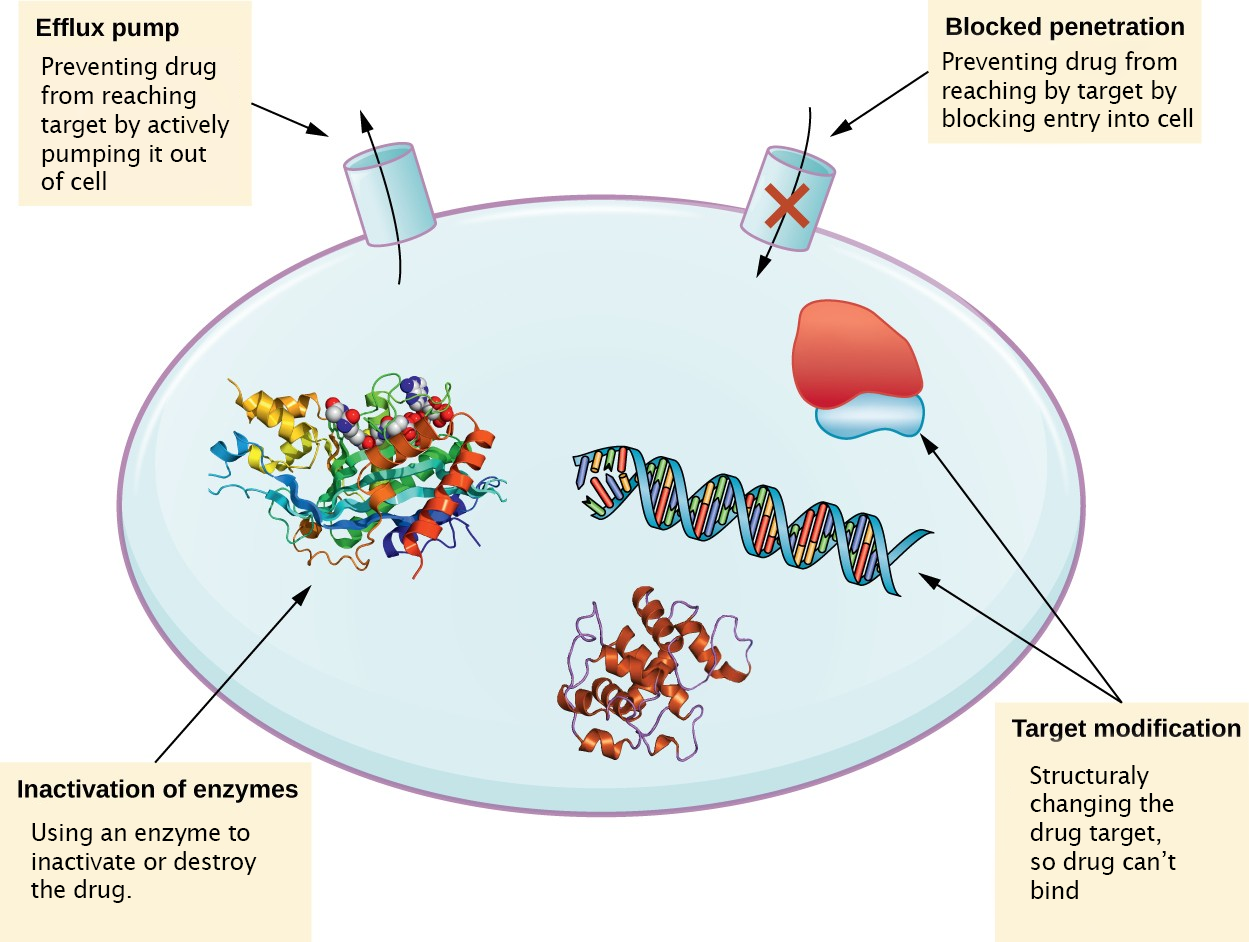
Drug Inactivation
Resistance genes may code for enzymes that chemically modify an antimicrobial, thereby inactivating it, or destroy an antimicrobial through hydrolysis. Resistance to many types of antimicrobials occurs through this mechanism.
Prevention of Cellular Uptake or Efflux
Microbes may develop resistance mechanisms that involve inhibiting the accumulation of an antimicrobial drug, which then prevents the drug from reaching its cellular target. This strategy is common among gram-negative pathogens and can involve changes in outer membrane lipid composition, porin channel selectivity, and/or porin channel concentrations. Additionally, many gram-positive and gram-negative pathogenic bacteria produce efflux pumps that actively transport an antimicrobial drug out of the cell and prevent the accumulation of drug to a level that would be antibacterial.
Target Modification
Because antimicrobial drugs have very specific targets, structural changes to those targets can prevent drug binding, rendering the drug ineffective. Through spontaneous mutations in the genes encoding antibacterial drug targets, bacteria have an evolutionary advantage that allows them to develop resistance to drugs. This mechanism of resistance development is quite common. Examples of this resistance strategy include alterations in
- penicillin-binding proteins (PBPs), providing resistance to penicillins
- ribosome subunits, providing resistance to macrolides, tetracyclines, and aminoglycosides;
- lipopolysaccharide (LPS) structure, providing resistance to polymyxins;
- RNA polymerase, providing resistance to rifampin;
- DNA gyrase, providing resistance to fluoroquinolones;
- metabolic enzymes, providing resistance to sulfa drugs, sulfones, and trimethoprim
- peptidoglycan subunit peptide chains, providing resistance to glycopeptides.
Target Overproduction or Enzymatic Bypass
When an antimicrobial drug functions as an antimetabolite, targeting a specific enzyme to inhibit its activity, there are additional ways that microbial resistance may occur. First, the microbe may overproduce the target enzyme such that there is a sufficient amount of antimicrobial-free enzyme to carry out the proper enzymatic reaction. Second, the bacterial cell may develop a bypass that circumvents the need for the functional target enzyme. Both of these strategies have been found as mechanisms of sulfonamide resistance.
![]()
- List several mechanisms for drug resistance.
Multidrug-Resistant Microbes and Cross Resistance
From a clinical perspective, our greatest concerns are multidrug-resistant microbes (MDRs) and cross resistance. MDRs are colloquially known as “superbugs” and carry one or more resistance mechanism(s), making them resistant to multiple antimicrobials. In cross-resistance, a single resistance mechanism confers resistance to multiple antimicrobial drugs. For example, having an efflux pump that can export multiple antimicrobial drugs is a common way for microbes to be resistant to multiple drugs by using a single resistance mechanism.
Methicillin-Resistant Staphylococcus aureus (MRSA)
Methicillin, a semisynthetic penicillin, was designed to resist inactivation by β-lactamases. Unfortunately, soon after the introduction of methicillin to clinical practice, methicillin-resistant strains of S. aureus appeared and started to spread. The mechanism of resistance, acquisition of a new low-affinity PBP, provided S. aureus with resistance to all available β-lactams. Strains of methicillin-resistant S. aureus (MRSA) are widespread opportunistic pathogens and a particular concern for skin and other wound infections, but may also cause pneumonia and septicemia. Although originally a problem in health-care settings (hospital-acquired MRSA [HA-MRSA]), MRSA infections are now also acquired through contact with contaminated members of the general public, called community-associated MRSA (CA-MRSA). Approximately one-third of the population carries S. aureus as a member of their normal nasal microbiota without illness, and about 6% of these strains are methicillin resistant.[1][2]
Multidrug-Resistant Mycobacterium tuberculosis
The emergence of multidrug-resistant Mycobacterium tuberculosis (MDR-TB) and extensively drug-resistant Mycobacterium tuberculosis (XDR-TB) is also of significant global concern. MDR-TB strains are resistant to both rifampin and isoniazid, the drug combination typically prescribed for treatment of tuberculosis. XDR-TB strains are additionally resistant to any fluoroquinolone and at least one of three other drugs (amikacin, kanamycin, or capreomycin) used as a second line of treatment, leaving these patients very few treatment options. Both types of pathogens are particularly problematic in immunocompromised persons, including those suffering from HIV infection. The development of resistance in these strains often results from the incorrect use of antimicrobials for tuberculosis treatment, selecting for resistance.
![]()
- How does drug resistance lead to superbugs?
Link to Learning
To learn more about the top 18 drug-resistant threats to the US, visit the CDC’s website (https://openstax.org/l/22CDC18drugres).
- A.S. Kalokhe et al. “Multidrug-Resistant Tuberculosis Drug Susceptibility and Molecular Diagnostic Testing: A Review of the Literature. American Journal of the Medical Sciences 345 no. 2 (2013):143–148. ↵
- Centers for Disease Control and Prevention. “Methicillin-Resistant Staphylococcus aureus (MRSA): General Information About MRSA in the Community.” http://www.cdc.gov/mrsa/community/index.html. Accessed June 2, 2016 ↵

