4 The Endomembrane System
Introduction
Since the authors of this textbook both did research in the area of endomembrane biology, we must admit that we are somewhat partial to this topic. That being said, the endomembrane system is arguably the most interesting and complex of all of the cellular structures we cover. It’s also quite challenging to study, as its function is essential to cell survival. Anything in the cell that is “essential” is also difficult to study, as the cell dies quickly if that function is disrupted.
There are several membrane-bound organelles that work together to form the endomembrane system: the endoplasmic reticulum (or ER for short), the Golgi apparatus (named after a person, so its name is capitalized), endosomes, lysosomes (called vacuoles in some taxa), and the vesicles that traffic cargo from one organelle to another. This system is extensive, and it is somewhat messy to differentiate one part from another. In this chapter we do our best to create order from the chaos and highlight how we study the endomembrane system as well as what we don’t yet know.
Topic 4.1: The Endoplasmic Reticulum
Learning Goals
- Explain the structural and functional relationships between the different compartments of the endomembrane system and identify them on different micrographs.
- Describe how proteins are targeted and imported into the endoplasmic reticulum and compare these mechanisms to protein targeting and import into the nucleus.
- Predict the targeting sequences required to insert a protein into the endoplasmic reticulum (ER) membrane in any orientation and predict protein topology from a corresponding domain map.
- Discuss the role of chaperones in protein folding and the role of the proteasome in the Unfolded Protein Response.
Introduction to the Endomembrane System
The endomembrane system consists of the endoplasmic reticulum (ER), Golgi apparatus, lysosomes, and endosomes. These compartments are involved in a great many cellular functions, including the processing of proteins that are destined for export from the cell, dealing with proteins that have been brought in from the outside of the cell, lipid synthesis, and a variety of signaling events.
The endomembrane system has a number of compartments, and cargo travels from one compartment to the next using smaller, membrane-bound structures known as vesicles. We say that the “start” of the endomembrane system is the ER, as this is the point of entry for newly synthesized proteins (Figure 04-01). Once proteins enter the ER they never return to the cytosol; they are carried by vesicle transport to the other compartments of the system. This flow of vesicles is highly regulated. The “end” of the endomembrane is usually considered to be the cell exterior, as proteins that pass through the entire length of this system, without being diverted, will eventually undergo secretion, ending up in the extracellular space, or embedded in the plasma membrane.
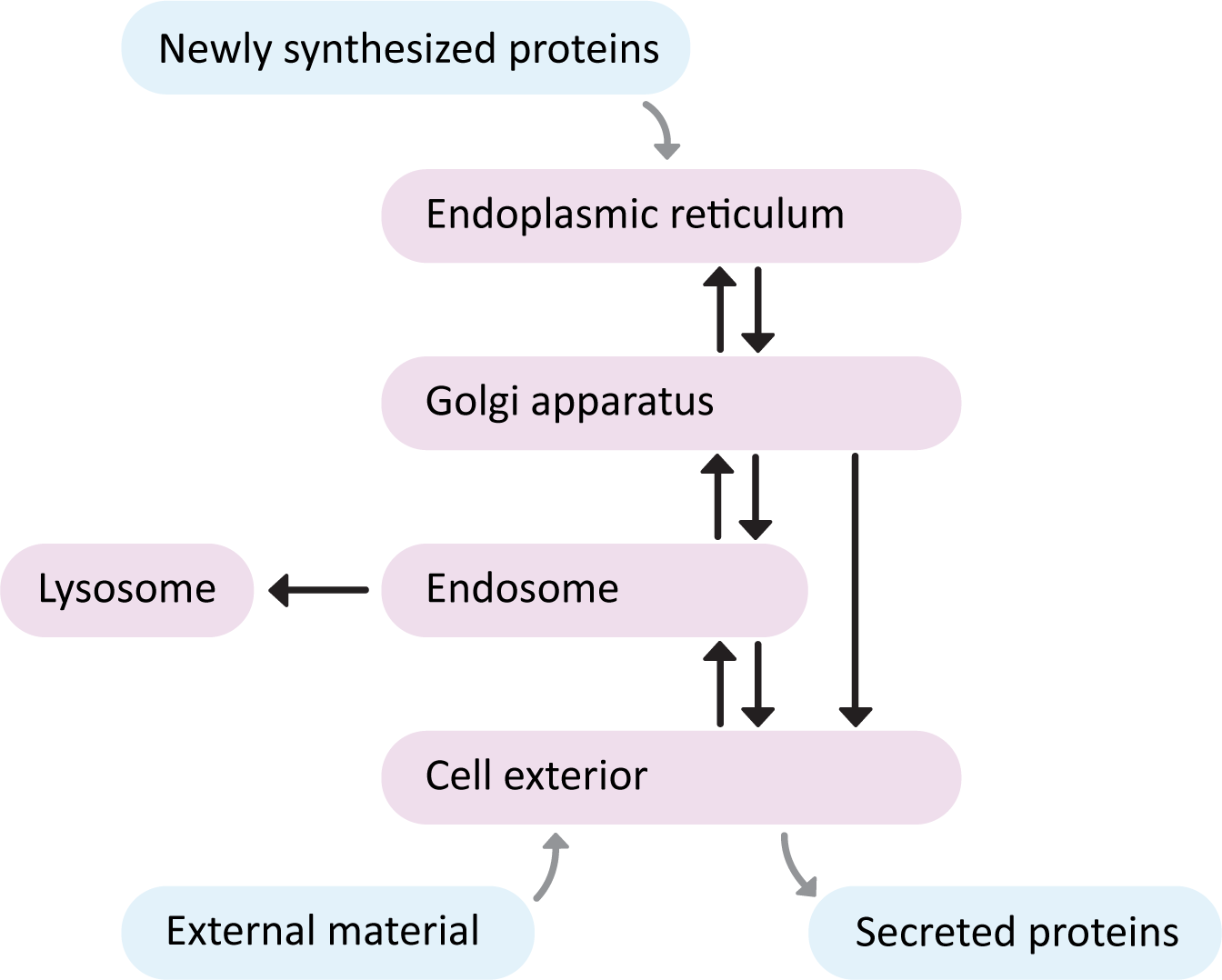
There are three major pathways to travel through the endomembrane system. See if you can trace each one in Figure 04-01.
- The secretory pathway, which is also often called the “default pathway” through the organelles. It is the path that most newly synthesized ER-targeted proteins will take as they eventually exit the cell. Proteins start in the ER, move to the Golgi and then finally moved to the plasma membrane.
- The lysosomal pathway, which is the path that newly synthesized digestive enzymes will take as they move to their eventual destination (the lysosome). Proteins start in ER, transported to the Golgi, delivered to the endosomes when they finally go to the lysosome.
- The endocytic pathway, is the path inward from the cellular exterior for proteins and substances that are unable to cross the membrane through diffusion or by transport protein. Material brought in via endocytosis will travel to endosomes and likely end up in the lysosome, where they can be degraded, and their components recycled into building block molecules. Molecules entering the cell this way are said to have undergone endocytosis.
Proteins that enter this interconnected set of organelles can only enter at the “start” of the pathway. So newly synthesized proteins that are destined for the secretory or lysosomal pathways must always enter at the ER. Material coming into the cell from the extracellular space via endocytosis begins at the plasma membrane. Additionally, proteins that enter the endomembrane system always travel the same route. They don’t skip compartments, or return to the cytosol (unless they are misfolded and need to be destroyed…but more on that later…).
In this topic, we have two main ideas to discuss:
- How proteins are modified after translation. This is called protein processing.
- Since protein processing begins in the cytosol and (sometimes) ends in the endomembrane system, we’re going to talk about this first, and then get into the details of how the endomembrane system works.
- How newly synthesized proteins enter the ER, thus gaining access to the rest of the endomembrane system.
Protein Processing in the Endomembrane System
As you (hopefully!) remember, protein translation happens in the cytosol. (As always, the introduction is there for you to review should you need it.) Once translated, all proteins are processed. Processing can include any number of events, such as the following:
- Folding: All proteins are folded to generate their 3D structure; thus, all proteins are processed in at least this one way. They will form the three (or four) levels of protein structure we mentioned in Chapter 2. The final folded 3D structure is based on the primary sequence of the protein.
- Removal of the first methionine at the N-terminus. Remember that the codon that serves as the “start” codon is also the codon that codes for methionine. In many proteins this methionine is removed once the protein is translated.
- Methylation
- Phosphorylation
- Acetylation
- Formation of disulfide bridges (most common in secreted proteins)
- Glycosylation (only in proteins destined for the cell exterior or the lysosome)
- Cleavage and more…
We will discuss some of these in more detail, but not all. We will focus first on protein folding and how it is managed by the cell. Then we’ll look briefly at how and where disulfide bridges form. Later in the chapter, we’ll look at how proteins are glycosylated, since that process is primarily the job of the Golgi apparatus.
Protein Folding
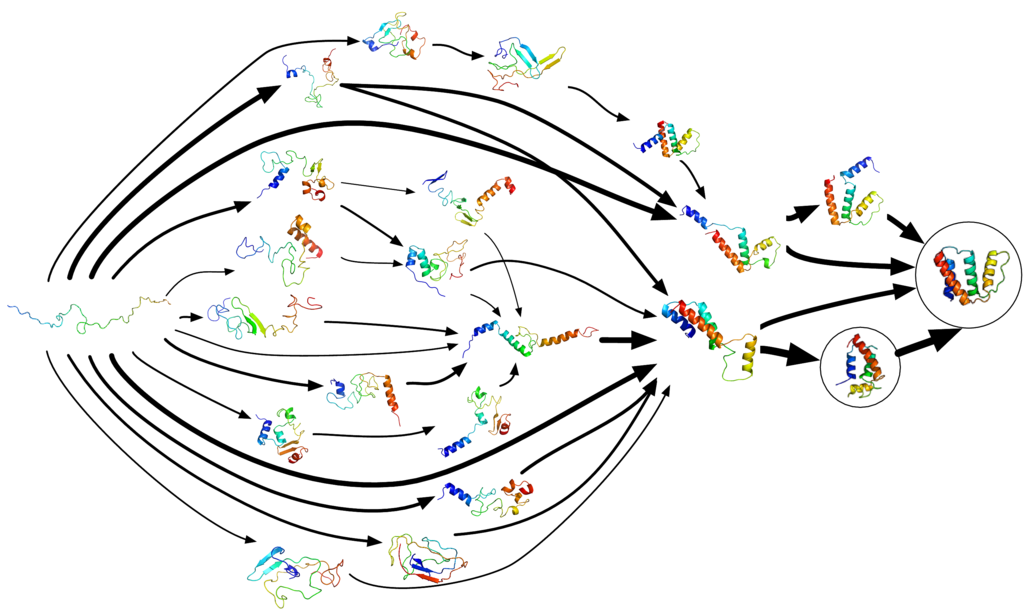
As the newly synthesized polypeptide emerges from the ribosome, it will immediately begin to fold based on its primary structure. Protein folding is a spontaneous process in which the polypeptide will take on the lowest energy conformation possible. This you should already know from our discussion of membrane proteins in Chapter 2. This is not to say that the protein will spontaneously “find” the proper conformation. Folding is extremely complex, and we continue to learn about it. What we do know boils down to this:
- Proteins can successfully fold a lot of different ways. They do not follow the same path each time.
- It is a process that can go wrong very easily. Not every translated protein ends up properly folded. Misfolded proteins are usually destroyed by the cell.
Figure 04-02, below, perfectly illustrates this concept. In it, we see many of the different steps a protein can take to get from “unfolded” to “properly folded.” Note that this image excludes all of the ways that a protein could fold and end up at a “dead end,” where it would not be able to find the appropriate final conformation. There are just as many, if not more, ways a protein can misfold. Since misfolded proteins are destroyed by the cell, it is unclear exactly how often it happens, but some estimates suggest that as many as 50% of proteins misfold!
Chaperone Proteins Help Ensure Proteins Fold Properly
The capacity for a protein to spontaneously fold correctly is important for more reasons than you might expect. Improperly folded proteins can be very detrimental to the cell. Not only would the proteins be unable to perform their function, but they might be insoluble and form large aggregates in the cell. In fact, misfolded proteins are a common source of disease, including Alzheimer’s, type 2 diabetes, cystic fibrosis, Parkinson’s, and prion diseases.
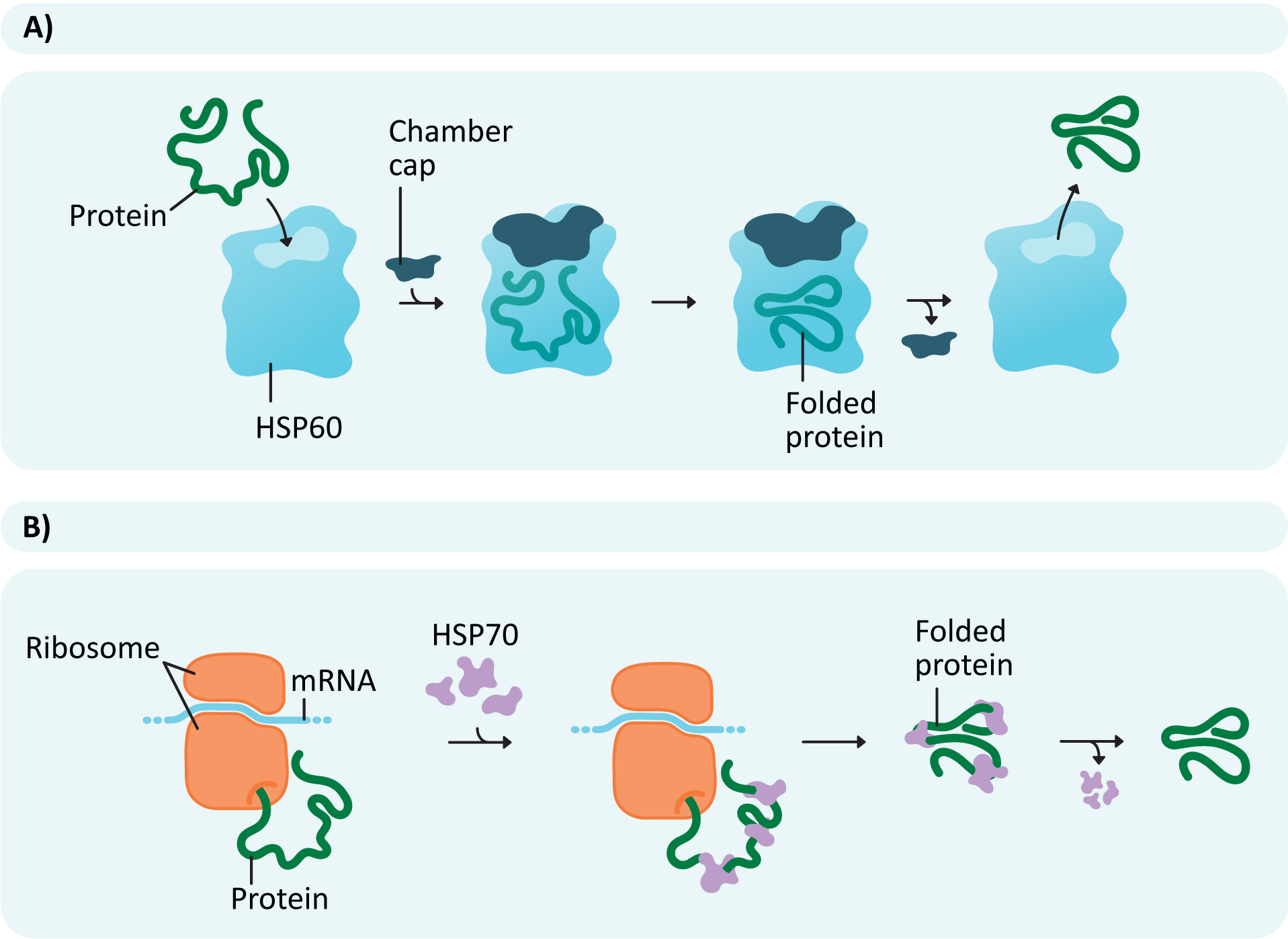
To ensure that the highest number of translated polypeptides manage to successfully fold, chaperone proteins often aid in the process (Figure 04-03 and Video 04-01). The new polypeptide forms a complex with chaperones that facilitate folding. Different chaperones help in different ways. Some may help simply by binding to specific regions of the polypeptide (Figure 04-03B) to prevent them from folding too early. This is how a chaperone known as HSP70 works. Others, such as HSP60 (or chaperonin), act as a chamber, providing a protected space away from the rest of the cytosol, where the protein can fold in isolation (Figure 04-03A).
Misfolded Proteins Are Sent to the Proteosome
It is vital that proteins either fold correctly or get disposed of properly when they misfold. The most obvious reason for this is that misfolded proteins are unlikely to function correctly, and this is true. However, it is equally important that these dysfunctional proteins get sent to the proteasome for degradation. The reason is that a misfolded protein is more likely to form complexes with other misfolded proteins nearby. This is due to exposed nonpolar regions on misfolded proteins that would otherwise be hidden away inside the protein. Misfolded proteins often form large clumps, or aggregates, inside the cell. The result of this is a giant blockage of protein inside the cell that gums up the works and inhibits function. If the aggregate is big enough or sticks around for a long time, it can actually kill the cell. There are a number of diseases in which protein aggregates accumulate in cells. For example, in Alzheimer’s disease and other forms of dementia, large protein aggregates are observed inside neuronal cells. One hypothesis is that these aggregates disrupt cellular function, and the cells die as a result. In sickle-cell anemia, the hemoglobin proteins misfold due to a mutation and form large clumps that ultimately destroy red blood cells.
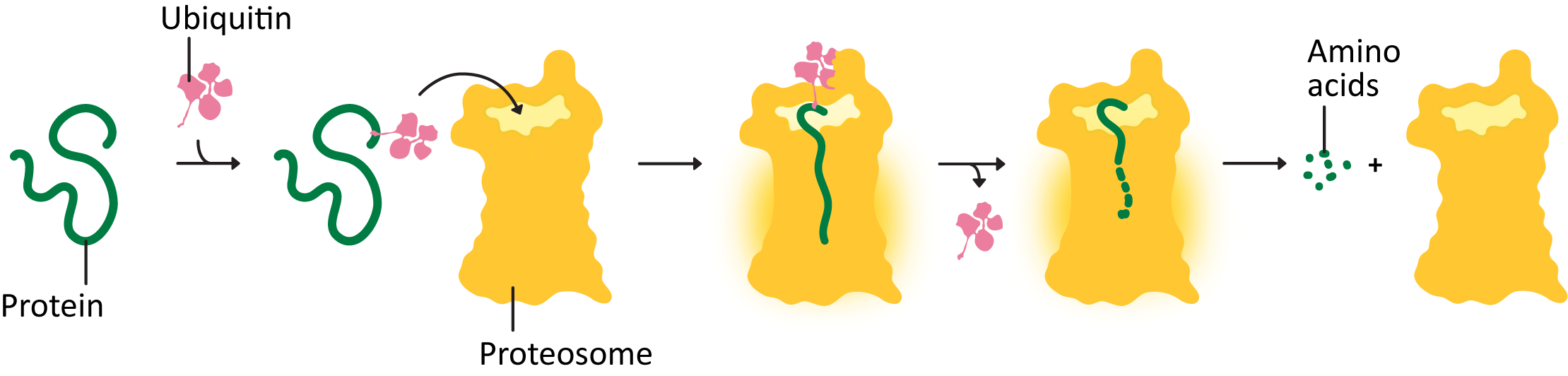
The proteosome is a large complex in the cytosol of the cell whose sole function is to destroy proteins that are damaged, misfolded, or no longer needed. It is one of the larger protein complexes inside the cell.
How does the proteosome work? Proteins that need to be degraded are tagged with a small peptide tag known as ubiquitin (Figure 04-04 and Video 04-02). The ubiquitin tag is recognized by other proteins, whose task is to deliver the ubiquitinated protein to the proteosome. Once there, the protein is threaded into the interior of the cylindrical center of the proteasome, and the peptide bonds are broken in a chemical process known as proteolysis. That way, the amino acids in the protein are kept and can be recycled by the cell. The ubiquitin is also removed and recycled.
Protein Folding and the Formation of Disulfide Bridges
Disulfide bridges are covalent bonds that form between the sulfurs of cysteines in an amino acid chain. Usually, the cysteines are far away from each other in the linear polypeptide chain but are brought together during folding. Enzymes usually facilitate the formation of disulfide bridges (also known as S-S bonds). These bonds help stabilize the 3D structure of a protein and/or help hold different subunits together. We touched on them briefly in Topic 2.3 as well, when discussing protein folding.
Interestingly, disulfide bridges only form under very specific conditions. They can only be formed under oxidizing conditions within the cell. (Hint: Remember your general chemistry on redox?) In reducing environments, they are unstable and tend to fall apart. What is the most interesting about this is that the cytosol tends to be a more reducing environment, which means that disulfide bridges are not easily formed or maintained there. On the other hand, the interior of the ER, known as the ER lumen, and the extracellular space tend to be oxidizing environments. This means that proteins that have moved through the ER during folding are far more likely to have disulfide bridges in them. We’ll see more on which proteins are folded in the ER and why a little bit later in this topic, but in a nutshell, proteins that are destined for any compartments of the endomembrane system or proteins that are targeted to the plasma membrane and/or the exterior of the cell are the ones that enter and do at least some of their folding in the ER.
The ER Is the Point of Entry into the Endomembrane System for Newly Synthesized Proteins
The ER consists of flattened membrane sacs, known as cisternae, and tubules. It is directly connected to the outer membrane of the nuclear envelope, but unlike most of the cartoons of the ER found in textbooks like this, it stretches throughout the entire cell. There is no part of the cell that is far from the ER. The ER has a number of different regions in it that are all connected together (Figure 04-05):
- Rough ER (rER). The cytosolic surface of the rER membranes has docked ribosomes that are synthesizing proteins for import into the ER. This is the site of synthesis for proteins destined for secretion, lysosomes, or membranes.
- Smooth ER (sER). This is continuous with rough ER, as shown in the photo below, and is the site of lipid and steroid synthesis. As mentioned in Chapter 2, new lipids and membranes are made in the sER.
- At the site where these two types of ER meet, there is a third form of ER known as transitional ER (tER). This is the site where vesicles usually form and newly synthesized proteins exit the ER in order to move on to the next destination (i.e., the Golgi apparatus). As such, this region is also known as an ER exit site.
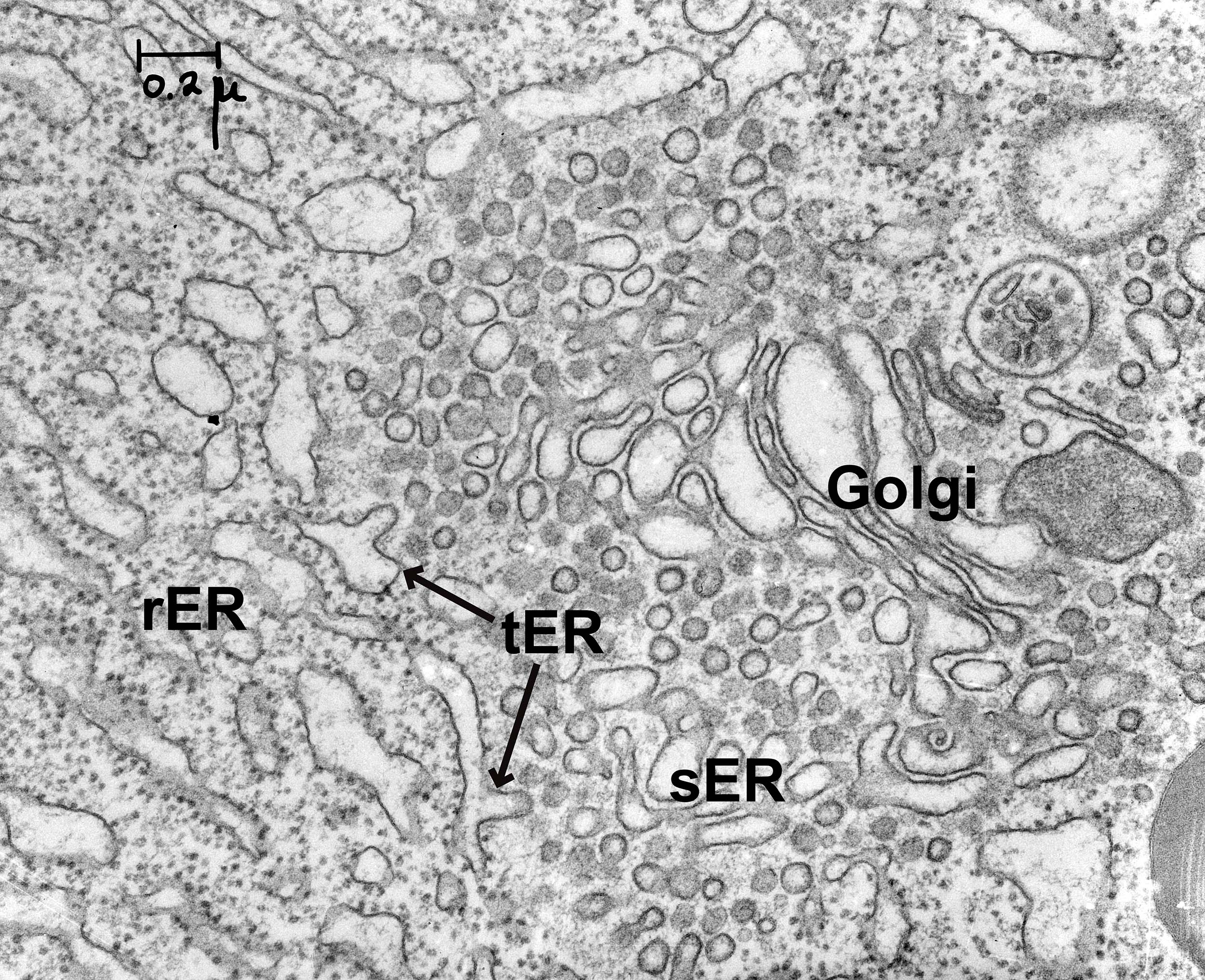
There are a couple of things to note about the traffic between the ER and the cytosol:
- The ER is the point of entry into the entire endomembrane system for newly synthesized proteins, so there is a lot of traffic heading from the cytosol to the ER.
- On the other hand, once a protein enters the ER, it does not return to the cytosol.
- Proteins that need to leave the ER to travel to the next compartment in the endomembrane system (which is the Golgi) will get packaged into vesicles, which will bud from the tER, and then fuse with the Golgi membrane. This way the protein can travel between compartments without returning to the cytosol.
Misfolded Proteins Leave the ER and Are Degraded in the Cytosol
There is one notable exception to the flow of protein traffic from the ER to the rest of the endomembrane system. Misfolded proteins that cannot be saved by the chaperone proteins must be sent to the proteasome for degradation. Since the proteasome is in the cytosol, this is the one instance when a protein will be transferred back across the ER membrane to the cytosol.
Just like in the cytosol, an accumulation of misfolded proteins in the ER lumen triggers the production of chaperone proteins and the expansion of ER, which can help reduce misfolding and aggregation. This is known as the unfolded protein response (UPR). Dealing with improperly folded proteins is thought to be a massive undertaking in the cell, which gets much worse when mutations exist that result in proteins that don’t fold efficiently.
Many proteins with multiple subunits, such as antibodies, are assembled in the ER. If these proteins are not properly assembled (e.g., via the formation of disulfide bridges), they will also trigger the UPR to address it.
Cells make lots of mistakes in the assembly of proteins. Proteins are made of hundreds, if not thousands, of amino acids, making them easy to misfold or misassemble. A huge part of the job of the ER is to ensure that properly folded proteins move on and misfolded ones are dealt with efficiently and don’t build up inside the cell and block traffic.
Entry/Exit from the ER Is Strictly Controlled
Just like the nucleus, the cell controls precisely what is allowed to enter and exit the ER. However, since the role of the ER is different from that of the nucleus and its structure far more intricate, controlling access is also more complex. Like the nucleus, only proteins with the proper targeting sequence will be allowed to enter the ER. Of those that are allowed to enter, some will become residents of the ER, while others will simply be passing through on their way to other destinations farther along in the secretory or lysosomal pathways.
Proteins Are Inserted into the ER Co-translationally
This is our second organelle in which we discuss protein import. Our first example was nuclear import (in Chapter 3). In the next chapter (Chapter 5), we will see how proteins enter the mitochondria and chloroplasts. In the case of the endomembrane system, there is more than one process to consider, as several organelles are included. Each compartment will have its own unique sequences, which must be properly read at the correct moment in the pathway. The additional sequences will be discussed later, when we explore the function of each of the compartments.
The first thing you must know is that translation of all proteins, including ER proteins, begins on free ribosomes in the cytosol (Figure 04-06). Whether or not a ribosome settles on the rER or stays in the cytosol to complete translation is dependent entirely on the protein it is currently translating. A ribosome that is currently translating a protein destined for the ER will attach itself to the surface of the rER during translation through a series of cues found in the amino acid sequence of the new protein. It is this field of ribosomes actively translating on the surface that gives the ER it its “rough” appearance. Smooth ER (sER) does not participate in protein synthesis (sER is important in lipid synthesis as well as metabolizing many toxic chemicals, like ethanol).
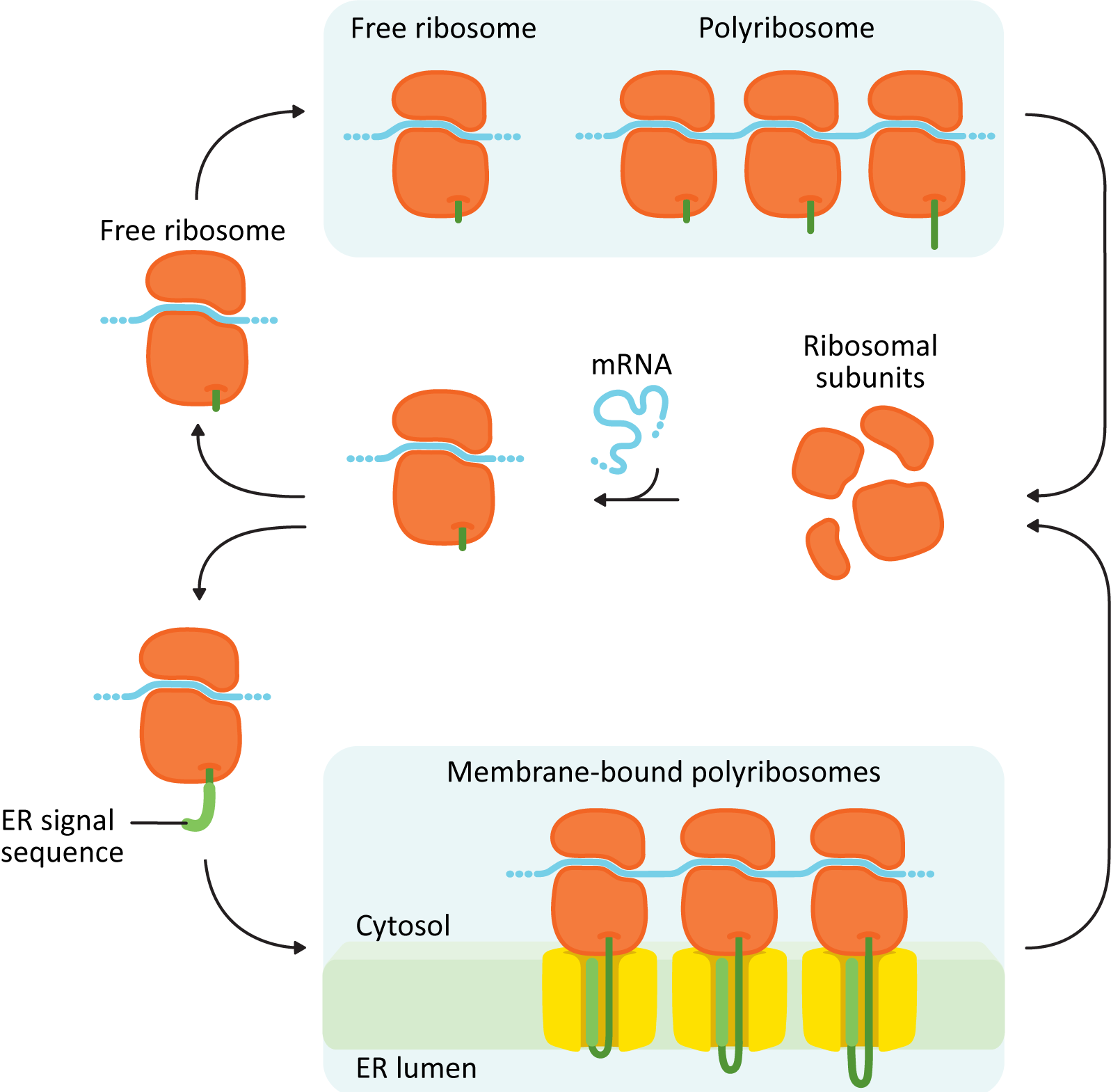
Structurally, the proteins entering the ER fall into two major categories:
- Soluble proteins: These proteins are completely translocated across the ER membrane into the ER lumen. They do not contain any membrane-bound portions.
- Integral membrane proteins: These proteins are only partially translocated into the ER and end up getting “stuck” with part of the protein embedded inside the membrane. These proteins may be destined for the ER, membranes of another organelle (Golgi, lysosomes, or endosomes), or the plasma membrane. Once a protein is inserted into a membrane, it cannot be removed.
The difference between the formation of a soluble protein and a membrane-bound protein is due to the number and placement of the ER insertion sequences. These sequences are used to identify when the ribosome should dock on the ER membrane and also which regions should become transmembrane domains. Here are some things to know about ER insertion sequences:
- Unlike the nuclear localization sequence (NLS), the specific order of amino acids in an insertion sequence is not as important as the chemical properties of the amino acids within the sequence. In all cases, the ER insertion sequence is about 8–10 nonpolar amino acids in a row.
- These ER insertion sequences go by a variety of names, depending once again on their location in the protein.
- If the sequence is directly at the N-terminus, it is called the signal sequence, the signal peptide, or an N-terminal START sequence.
- If the sequence is anywhere else within the primary sequence, it will be called an internal START or STOP transfer sequence, depending on how it aligns with the other ER sequences in the polypeptide.
- Despite the different names and different locations, the sequence itself remains the same. It is still 8–10 nonpolar amino acids, as mentioned above. The location of these targeting sequences within the polypeptide chain determines whether a protein is soluble or an integral membrane protein and also impacts the structure and orientations within the membrane.
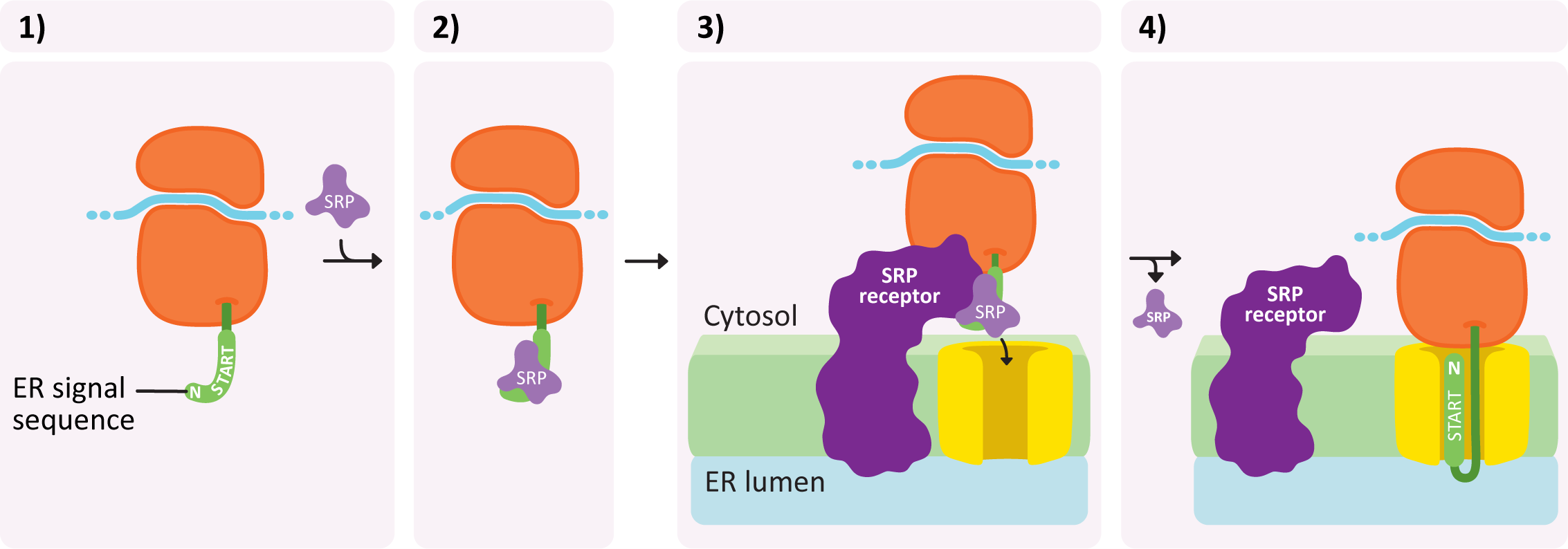
Insertion into the ER takes place in a number of steps and is illustrated in Figure 04-07:
- An ER insertion sequence is translated and almost immediately recognized by a ribonucleoprotein known as the signal recognition particle (or SRP).
- The SRP binds to this sequence and inhibits translation.
- The entire complex (ribosome + mRNA + partially translated protein) is brought to the ER and binds to a special SRP receptor protein in the ER membrane. The ribosome becomes attached to a translocation channel for the newly synthesized polypeptide. This attachment is facilitated by the SRP receptor and requires GTP as an energy source.
- As the ribosome becomes attached, the SRP is removed and translation resumes, but now the new protein is being pushed through the translocation channel into the ER lumen.
Video 04-03 does an excellent job of showing the ribosome docks onto the ER at the molecular level. However, it has no narration, so you’re going to have to consider when each step is happening on your own.
Different Types of Protein Insertion into the ER Membrane
Since both soluble and membrane-bound proteins are inserted in this way, using these same insertion sequences, it stands to reason that we need to explore a few different scenarios. In all cases, the location and order of the various ER insertion signals will determine whether a protein is soluble or membrane bound as well as how many times it passes through the membrane. Let’s explore how this works.
Soluble Proteins
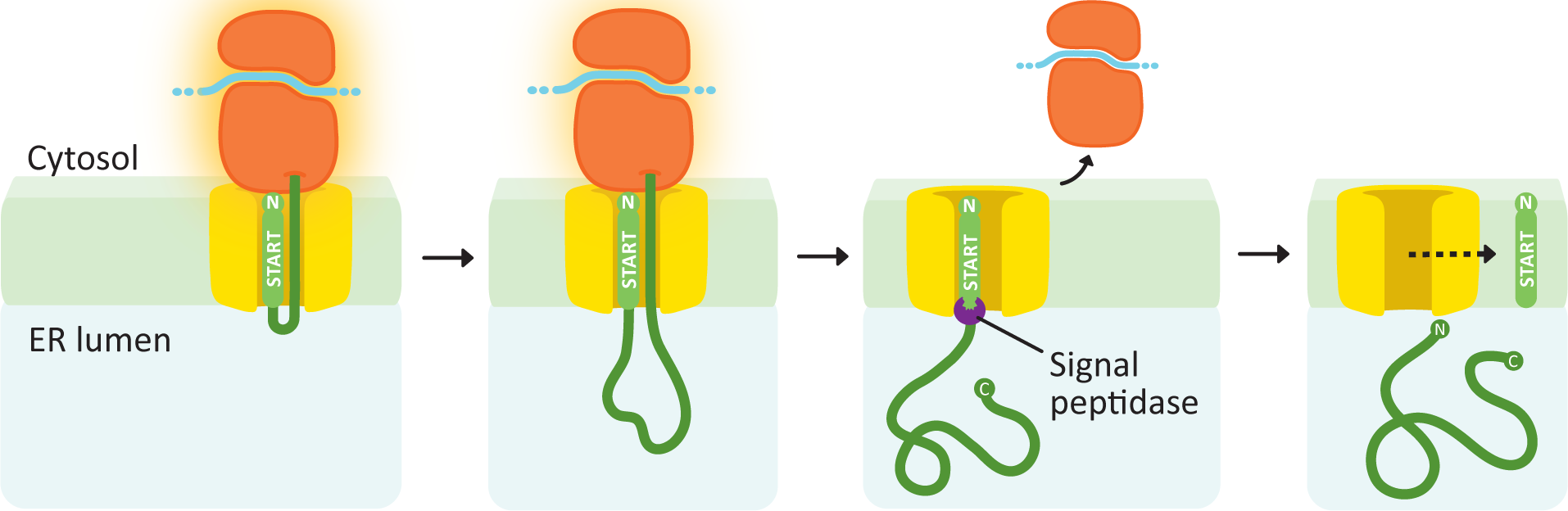
Soluble proteins need only a single transfer sequence, and it is always found at the N-terminus (Figure 04-08). After the signal sequence is recognized, the ribosome docks, and the polypeptide is threaded through the translocation channel into the lumen of the ER as it is synthesized. The signal sequence remains embedded in the membrane and is later cleaved off by a protein called the signal peptidase. Once that happens, the new protein is free and soluble in the ER lumen.
Membrane Proteins
A key point in the production of membrane proteins is that the orientation of a protein in the membrane is established when it is first inserted into the membrane, during translation. The orientation of the protein persists throughout its life-span, even as it is shuttled from one compartment to the next. That is, the cytosolic side of the protein remains on the cytosolic side of the membrane throughout its entire life. More on this later.
As membrane proteins are being translated, ribosome docking and co-translational insertion will not begin until an ER insertion sequence is encountered. Thus, the very first insertion sequence encountered is called the “START transfer” sequence. The first START may be at the N-terminus, as we saw in the previous example, but it doesn’t have to be.
The new protein will continue to be translated into the ER until a stop codon is reached (which ends translation) or a second insertion sequence is encountered. The second insertion sequence serves as a “STOP transfer” signal, which will close the translocation channel, release the ribosome, and stop co-translational insertion. When the START and STOP sequences are inside the amino acid sequence (i.e., not at the N-terminus), they serve as transmembrane domains for the growing protein. If the ribosome is still translating the protein after the STOP is encountered, it will remain tethered to the ER via the translating protein until either a new START sequence is met or translation ends. This process can be repeated many times in a single protein for as many ER insertion sequences as exist in the primary structure.
To further clarify how the ER insertion sequences are used to create membrane proteins, we have provided a number of examples.
Example #1
A single internal START sequence produces a protein with one transmembrane domain and the N-terminus on the cytosolic side.
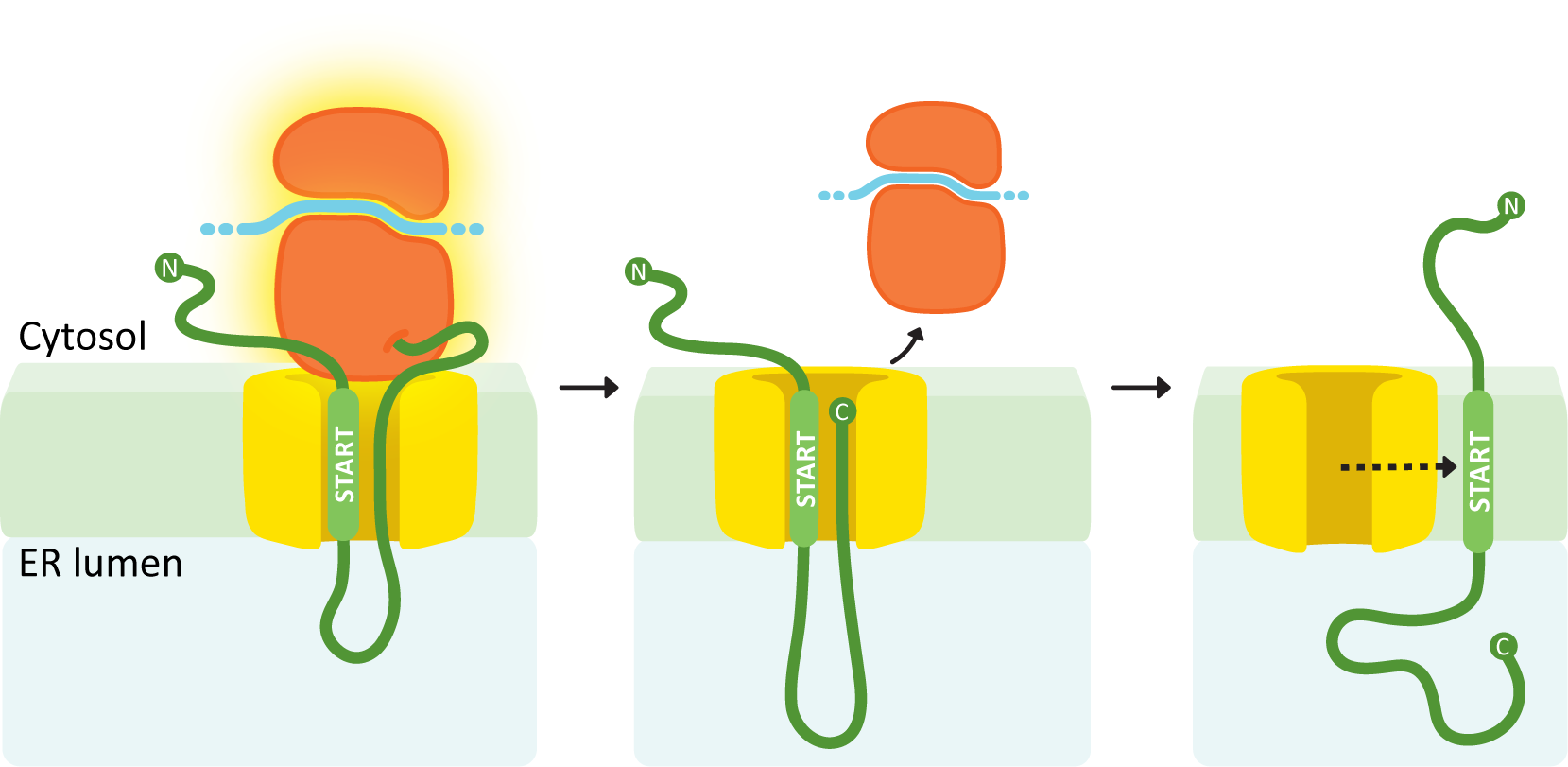
In this example (see Figure 04-09), the internal ER insertion sequence is recognized partway through protein synthesis, and the ribosome is brought to the translocation channel. That sequence is the START transfer, and everything after it is threaded through the translocation channel. Once translation is finished, the ribosome is released. The translocation channel opens and allows the protein to diffuse laterally into the membrane. The internal START transfer sequence is not cleaved and instead remains embedded in the membrane, becoming the transmembrane domain for the protein. As discussed in Chapter 2, the transmembrane domain of the protein holds the protein in the membrane because of the very strong association between the nonpolar amino acids in this region and the nonpolar lipid tails in the lipid bilayer. The N-terminus remains in the cytosol, while the C-terminus ends up inside the ER lumen.
Example #2
An N-terminal START transfer sequence followed by a STOP transfer creates a single-pass protein with an N-terminus in the lumen and a C-terminus in the cytosol.
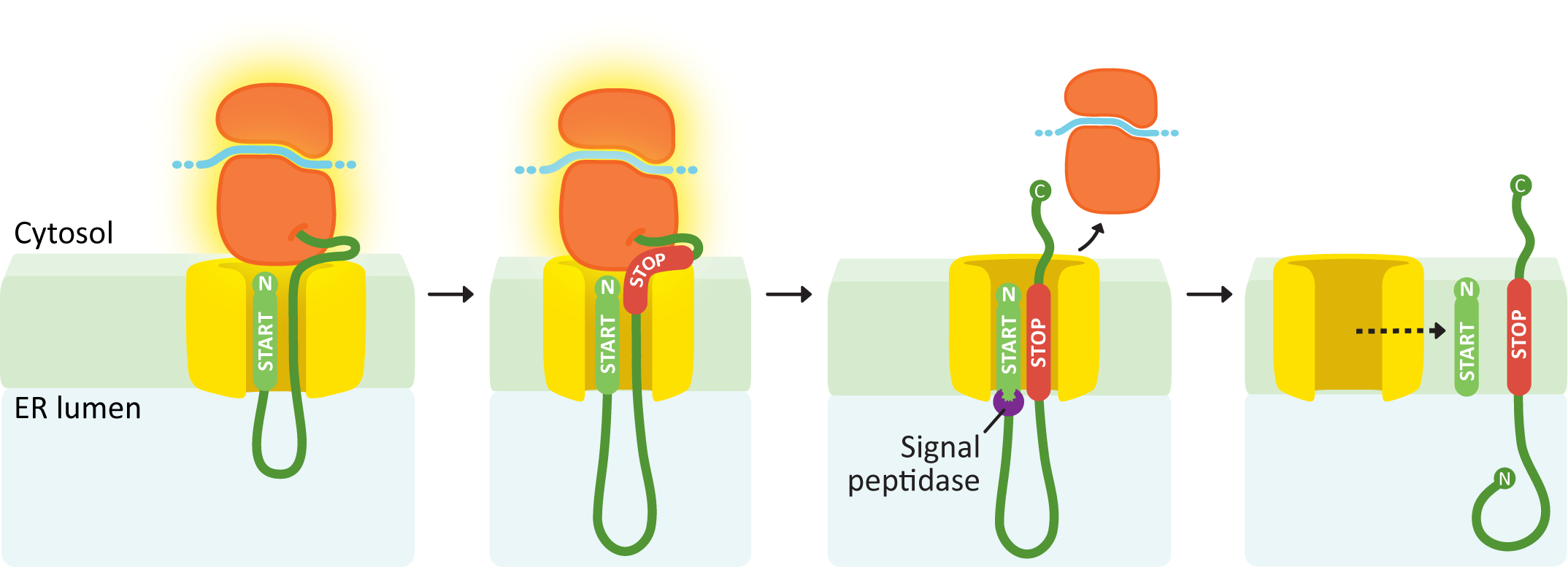
Similar to the soluble protein example (Figure 04-08), this next example protein has an N-terminal START sequence (Figure 04-10). This targets it immediately to the ER membrane, and the growing polypeptide is threaded through the channel from the very beginning of translation. At some point during translation, it runs into another ER insertion sequence, which will act as a STOP transfer sequence. When the STOP transfer sequence is encountered, it causes the translocation channel to stop threading polypeptide through the translocation channel into the ER lumen. The ribosome undocks from the membrane but is still tethered via the translating protein. Since there are no other insertion sequences in this protein, the ribosome will complete translation undocked but tethered. Since the START sequence is N-terminal, it will be cleaved off, which results in the N-terminus of the protein being free in the ER lumen, while the C-terminus remains in the cytosol.
Example #3
An internal START followed by a STOP creates a double pass membrane protein with both an N-terminus and a C-terminus in the cytosol.
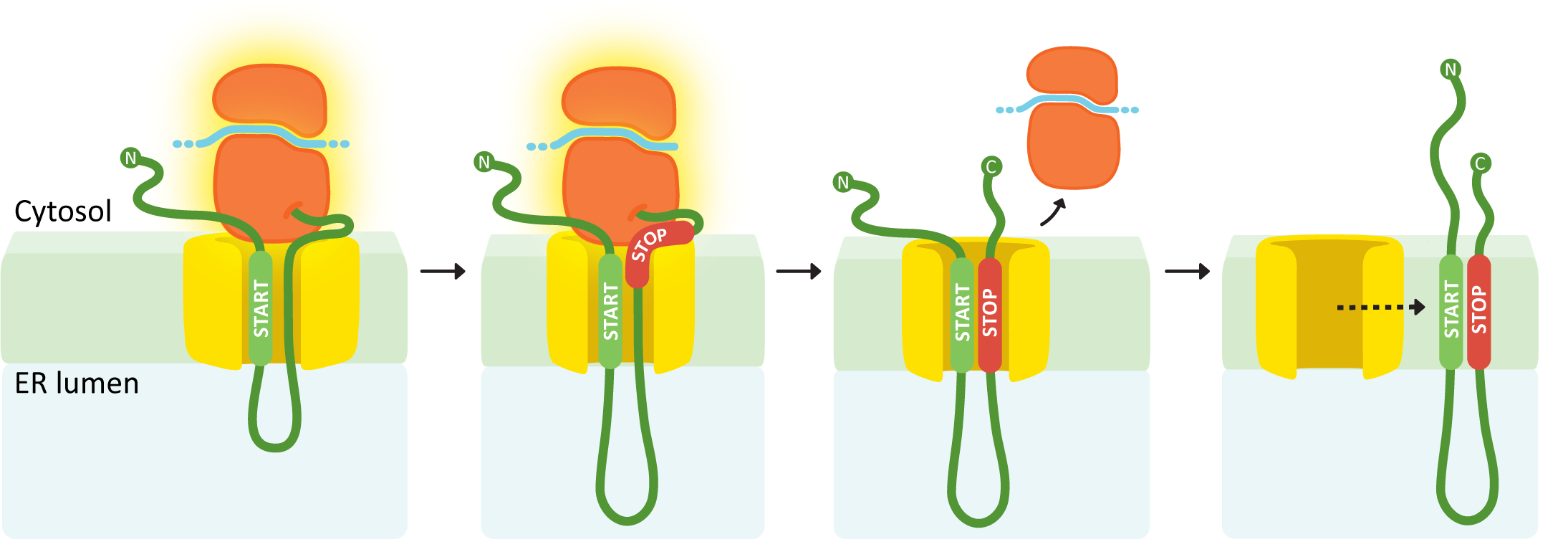
In this example (Figure 04-11), the first insertion sequence encountered is in the middle of the polypeptide, so it will be an internal START. Again, the internal START sequence initiates ribosome docking, and the polypeptide begins to thread through the translocation channel. This continues until the second insertion sequence is encountered (a STOP transfer sequence). The STOP sequence ends insertion through the translocation channel, and the ribosome once again completes translation undocked but tethered to the ER. Once translation is complete, the START and STOP are released from the translocation channel and diffuse laterally into the membrane. In this case, both insertion sequences are retained and become transmembrane domains. Both the N- and C-termini of the resulting protein will be in the cytosol.
Example #4
A protein with many membrane-spanning regions.
Using the three previous scenarios as a foundation, you can create a protein with any number of transmembrane domains simply by adding more insertion sequences, which become alternating START and STOP transfer sequences. A START transfer sequence will bring the ribosome to the translocation channel to thread the growing polypeptide into the lumen, and the STOP transfer sequence will release the ribosome, thereby ending the continued use of the translocation channel. The internal START and STOP transfer sequences will each become transmembrane domains, whereas an N-terminal START (if present) will get cleaved. In this way, the protein is essentially stitched into the membrane, and the ribosome is bound and released a number of times. Our final example (Figure 04-12) has six transmembrane regions with the N- and C-termini in the cytosol.
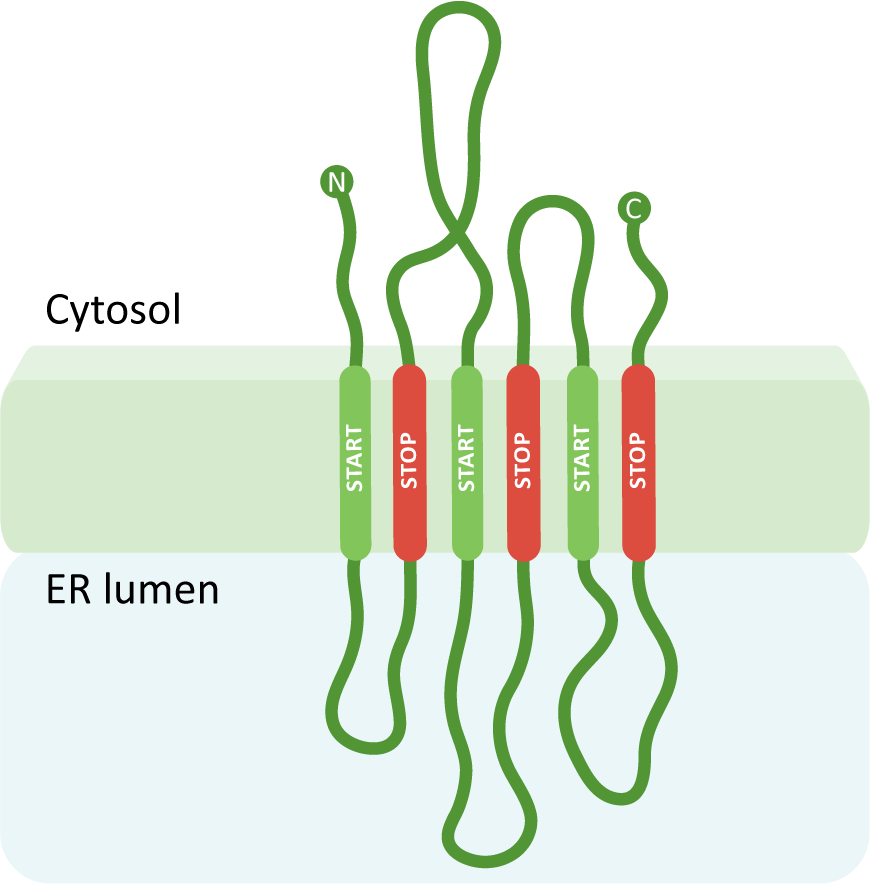
We hope that going through these examples has helped make sense of how protein ER insertion sequences work. To briefly summarize, we want you to take away the following the key points from this discussion of protein insertion into the ER:
- The ER insertion sequence is always 8–10 nonpolar amino acids. The difference between how each of the sequences is treated by the cell is due entirely to its location within the primary sequence of the growing polypeptide.
- There are two major categories of hydrophobic signals used in the insertion of membrane proteins. All of these are membrane-crossing domains:
- START transfer sequences. There are two kinds of start transfer sequences:
- N-terminal START transfer sequence. Like its name says, this sequence is at the N-terminus of the protein. It remains in the membrane during translation and is cleaved off of the protein by the signal peptidase. This is also called the signal peptide or signal sequence.
- Internal START transfer sequence. Similar to a signal sequence but located internally (i.e., not at the N-terminal end of the protein). It also binds to the SRP and initiates transfer into the ER. Unlike the signal sequence, it is not cleaved after transfer of the protein.
- STOP transfer sequence. A STOP transfer is never the only signal in a polypeptide chain. It follows either an N-terminal or an internal START transfer sequence. The STOP transfer signal is a membrane-crossing domain. It remains in the membrane. The sequence is not cleaved.
- START transfer sequences. There are two kinds of start transfer sequences:
- The orientation of the protein in the membrane is completely dependent on whether there is an N-terminal START sequence or not. If the protein has an N-terminal START, then the N-terminus of the protein will be in the ER lumen. If the first ER insertion sequence encountered is internal, then the N-terminus will remain on the cytosolic side of the ER membrane.
- The number of transmembrane domains a given protein will have will be equivalent to the number of internal transfer sequences (i.e., all STARTS and STOPS that are anywhere other than at the N-terminus).
Video 04-04 is an animation that does an excellent job of showing the details of protein insertion into the ER. We find that students really benefit from seeing this concept in action.
ER Resident Proteins Require an Additional Signal to Stay in the ER
ER resident proteins are proteins that are retained in the ER, as this is where they function. The chaperone proteins that are sometimes required inside the ER to help with protein folding are a great example of an ER resident. These proteins are inserted into the ER as usual, using the ER insertion sequences in their primary sequence, but also carry an ER retention signal called the KDEL (or HDEL in some species) sequence. It is named the KDEL sequence because this is the one-letter code for each of the amino acids in the sequence. It is located in the primary sequence of the protein, always at the C-terminus. The KDEL sequence ensures that any ER resident proteins that might accidentally get packaged into vesicles and shipped to the Golgi will be captured and sent back to the ER.
Protein Insertion and Membrane Asymmetry
As you may recall from Chapter 2, membranes are asymmetric, meaning that the cytosolic side is different from the noncytosolic side. Part of this membrane asymmetry is provided by the location and orientation of the proteins in the membrane. The initial insertion of these proteins into the ER membrane is a big part of how that asymmetry is established. Once that protein is inserted into the membrane, it can’t be removed, so it needs to be done right.
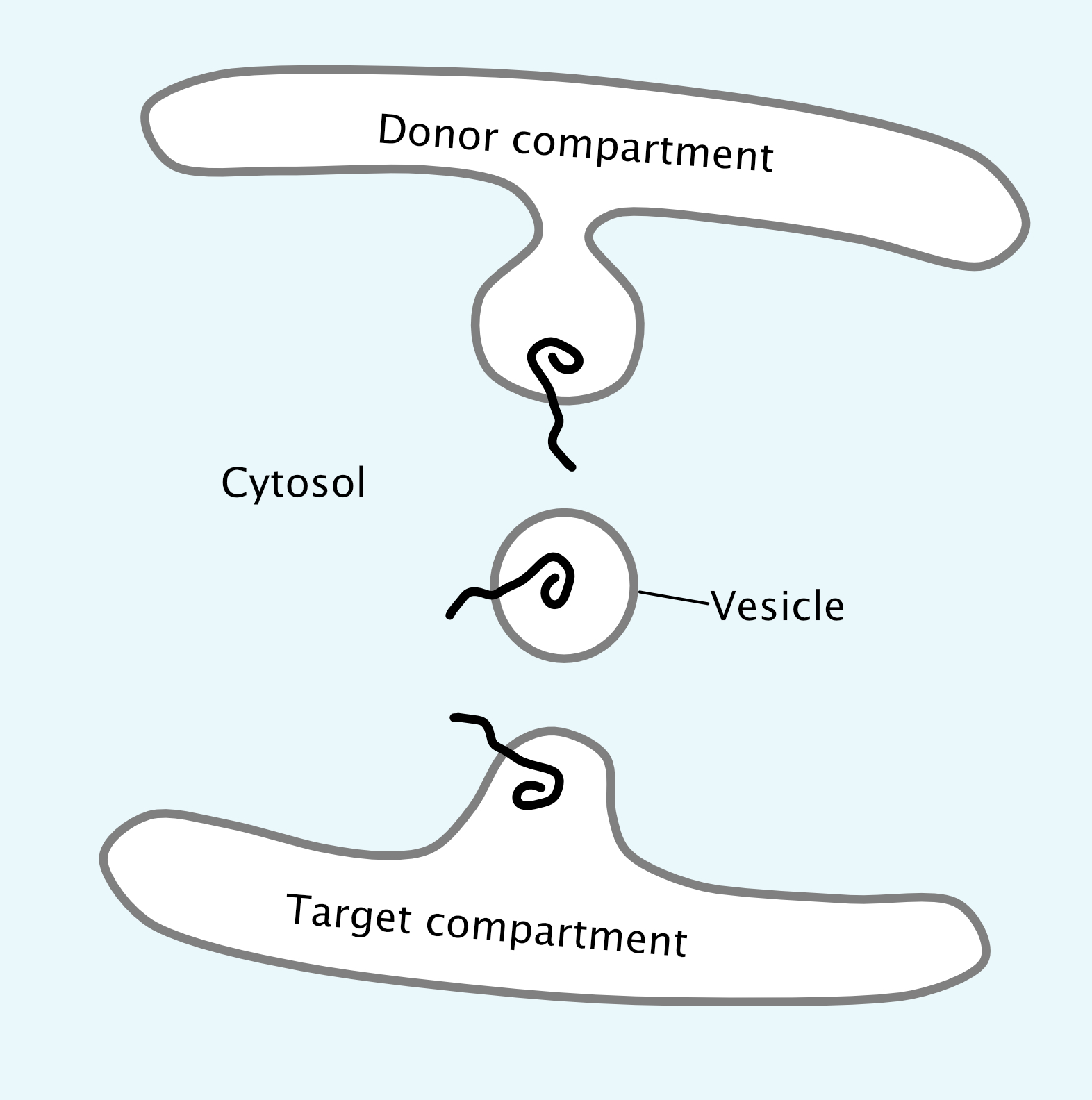
It is also very important that proteins remain properly oriented in the membrane as they move through the organelles of the endomembrane system toward their final destination. The use of membrane-bound vesicles is ideal for this, as the method of budding and fusion of vesicles maintains the orientation of the protein in the membrane while also eliminating any possibility of release into the cytosol (Figure 04-13). In all cases, and at every step, the cytosolic side of the membrane faces the cytosol. This is a key concept in understanding the endomembrane system. Check out the protein orientation in Figure 04-13 and notice that the part of the protein pointing into the cytosol is always the same part even when it gets to the donor compartment.
putting it into practice
Mapping Protein Domains
As we learned in Chapter 2, bioinformatics is a useful tool to identify known patterns within a protein sequence. We also explored how hydropathy plots allow us to identify potential transmembrane domains based on the amino acid sequence of the protein. If we add that to the information you’ve learned about so far in this chapter, we get a clearer picture of how we can learn about proteins using bioinformatics. You now know that when a hydropathy plot identifies hydrophobic sequences, it’s also identifying the ER insertion sequences that exist within the amino acid sequence. Hopefully, this shows you how the signatures of many functions of proteins exist in their amino acid sequences. Other sequences, such as the NLS or the KDEL, would also be easily recognized by an algorithm that was created to search for them. Import sequences are a great example, but they are by no means the only patterns or motifs that we can identify in a sequence that tell us something about protein function. Regions of a protein that have a specific function attributed to them are called protein domains.
With all of the different patterns and motifs within proteins, scientists need a system to keep track of which motifs exist in any given protein based on the bioinformatic analysis. Visual representations of the linear protein chain are used to map signature motifs within proteins. These are known as domain maps. Figure 04-14, below, shows how we could collect the hydropathy plot data from a given protein (Glycophorin A) and translate them into a domain map that shows how these regions function. Glycophorin A is a membrane protein found in red blood cells (see Figure 02-18, found in Topic 2.3).
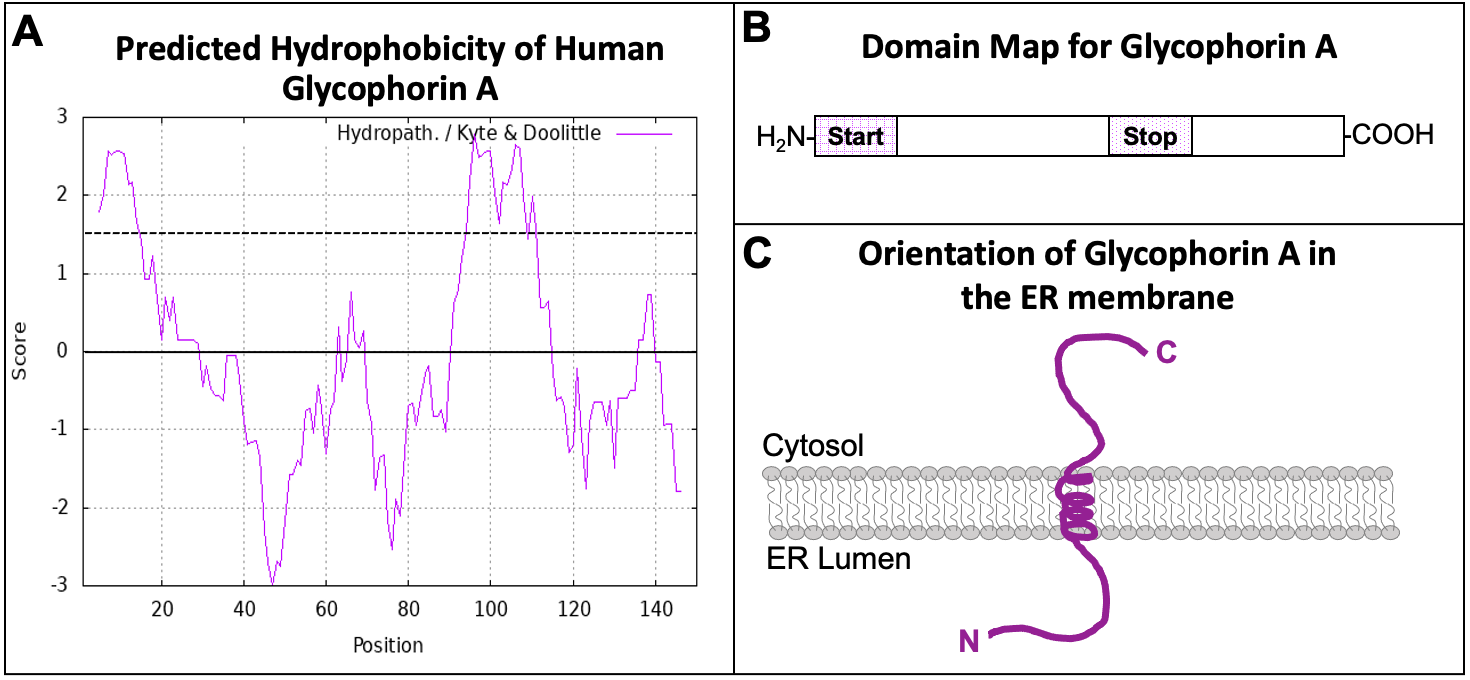
The hydropathy plot of Glycophorin A (Figure 04-14A) shows two regions within the amino acid sequence that have high hydrophobicity. These two regions are the two ER insertion sequences. Using the information in this hydropathy plot, we can draw a domain map for the protein (Figure 04-14B). The first hydrophobic region is directly at the N-terminus, which makes it an N-terminal START sequence. The second is in the interior of the protein, and it follows a START, so it is a STOP transfer sequence, and it will become a transmembrane domain. From the domain map that we have created, we can also draw the predicted protein and its orientation in the ER membrane (Figure 04-14C). Since the orientation of the protein is preserved as it moves through the endomembrane system (Figure 04-13), you should also be able to predict the orientation of the protein in its final destination, which would be the plasma membrane in the case of Glycophorin A. The portion of the protein that is currently in the ER lumen will eventually be placed on the exterior side of the plasma membrane. The cytosolic side of the protein always remains on the cytosolic side.
For this example, we have focused specifically on mapping ER insertion sequences. However, it is important to remember that there are lots of things that can be mapped in this way. For example, you can map all of the different localization sequences (i.e., NLS, NES, ER insertion, KDEL, etc.) onto domain maps. If the protein was an enzyme with an active site that could be identified, then that could also be mapped. Many secreted proteins also have sites where they get glycosylated. Glycophorin A (the example from Figure 04-14) is glycosylated multiple times. If you were to follow the link to the bioinformatic protein data, the glycosylation sites are also identified and mapped. Maps of this kind are very important for helping us visualize and understand the structure and function of the proteins we study. You will likely have the opportunity to practice domain mapping as we continue to work through the various organelles and how proteins are targeted to them.
Topic 4.2: Vesicle Transport
Learning Goals
- List the four stages of vesicle transport, and list the protein machinery involved at each step.
- Explain how vesicle coats facilitate cargo loading and vesicle budding.
- Discuss the importance of maintaining specificity in both docking and fusion of vesicles at their target compartments and how Rabs, tethers, and SNAREs facilitate this process.
Introduction
As we mentioned in the previous topic, the ER is the point of entry for all newly synthesized proteins that either reside in the endomembrane system or are destined to be secreted. In addition, once proteins enter the endomembrane system, they do not return to the cytosol. This poses a unique challenge for the cell in that the organelles of the endomembrane system are not physically connected to each other, and yet they must transfer cargo from one organelle to the next without allowing cargo to “escape” back to the cytosol. As such, vesicles are used to shuttle cargo around the cell.
Vesicles carrying cargo bud from one of the membrane-bound organelles of the endomembrane system (ER, Golgi, endosome, lysosome, etc.). They then move through the cytosol to fuse with the next organelle in the endomembrane system or the plasma membrane (Figure 04-15). In this topic, we will examine the details of how vesicles form, travel, and then fuse with the membrane at their target destination.
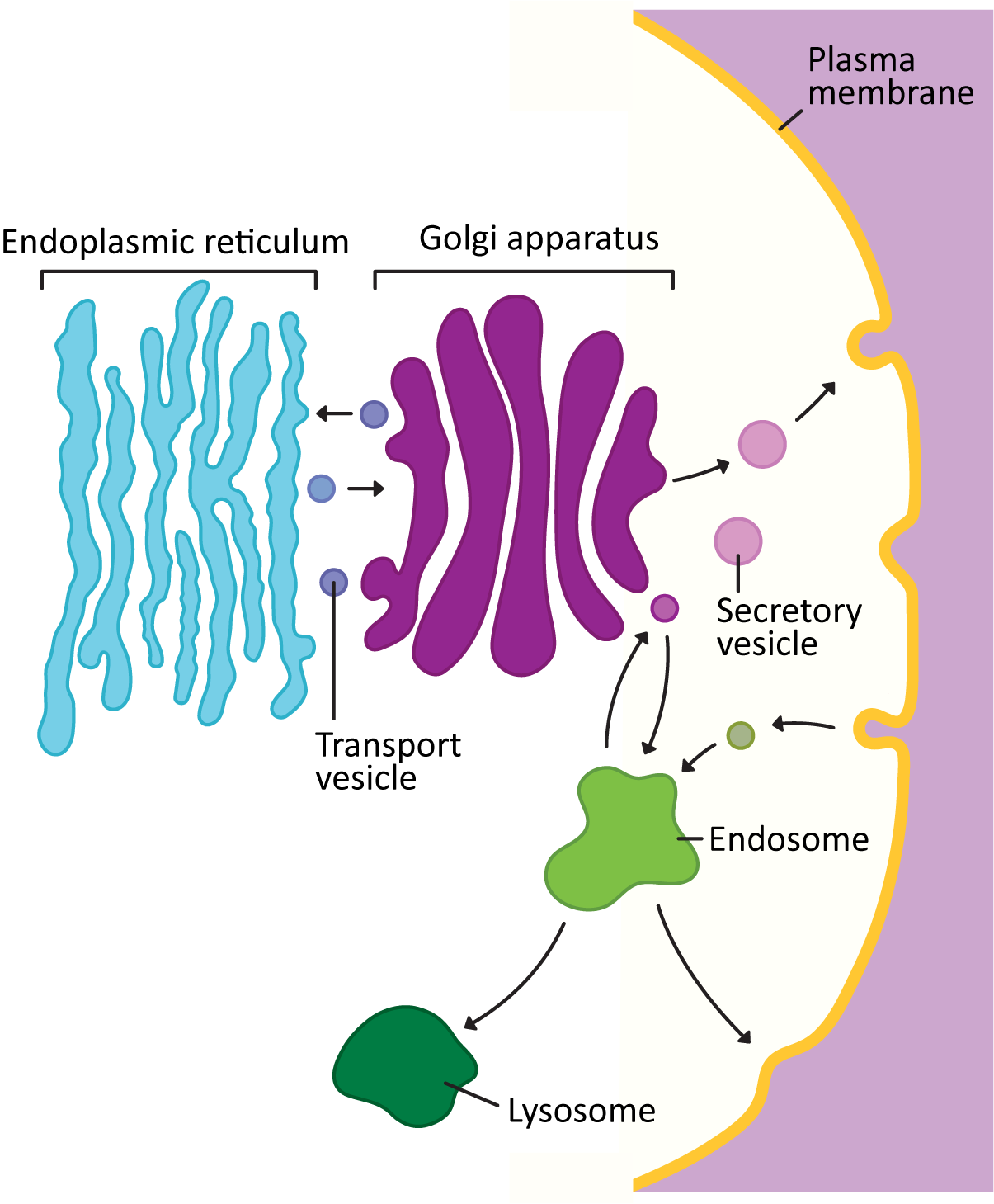
It’s important here for us to take a moment and point out that vesicle trafficking is extremely complex! There’s a lot we know about how vesicle trafficking works, but there’s also a lot that we don’t know. Vesicles are produced by all of the organelles of the endomembrane system. They all have specific cargoes and specific destinations. Some organelles (like the Golgi) are producing separate vesicles that will be sent to any number of different locations. The cargo is sometimes quite large, making it very difficult to put in a vesicle (collagen is a great example). And yet it, too, must get packaged properly and sent to its destination. For each new insight we gain about vesicle transport, we also find out that there are exceptions to what we thought were “the rules.” This situation can become overwhelmingly complex if we also consider all of the specialized cell and tissue types, differences between organisms, and even differences between biological kingdoms and how their endomembrane systems have evolved to address their unique needs.
Despite the complexity, in this section, we provide you with a framework to understand the parts of vesicle traffic that apply in most situations. This will hopefully help you learn what to look for when encountering a new trafficking scenario.
All of these pathways through the endomembrane system rely heavily on vesicles to transport their cargo from one stage in the pathway to the next. Since the three major pathways (see Figure 04-01 to remind you of what they are) are also always running in tandem, we begin to see why traffic through the endomembrane system is so complex.
Vesicle Traffic
The Basics
The basic principle of vesicle trafficking is that vesicles must take the correct cargo from the donor compartment and deliver it to the correct target compartment. This means that
-
-
- the correct cargo must get into the correct vesicle, and
- the correct vesicle must get sent to the correct destination.
-
A good analogy to consider here is that of an Amazon order (or any other online/mail order). When an online order is placed, we expect a couple of things to happen. The first one is that the product that was ordered will be properly packaged with the rest of the things that were ordered and prepared for shipping. This implies that no one else’s order will be included in this package. The second is that the package will then be delivered to the correct address, which is identified by the original order form. If you have ever had an online order that was shipped incorrectly, you have an understanding of the importance of doing this correctly the first time.
Vesicle trafficking works in a similar way. Instead of mailing addresses, targeting sequences are used. These sequences determine which type of vesicle the protein will enter so that it can be sent to the correct destination. The exception is when the cargo is destined to be secreted via the secretory pathway. Since secretion is considered the “default pathway” of the endomembrane system, proteins that are in the ER that don’t have other targeting sequences will get scooped up into vesicles in a nonspecific way and sent along on their journey. We’ll see more about the different pathways through the ER later in this chapter.
Vesicle traffic is controlled by the protein machinery that helps the vesicle form, travel, and fuse with the target membrane. It is this protein machinery that is the focus of this topic.
In all vesicle traffic, there are four main stages to the process (see Figure 04-16):
It is sometimes said that there is a fifth stage, which is resetting the system so that the next round of trafficking can happen. Repeated rounds of vesicle traffic require that the trafficking machinery gets recycled back to its original location so that new vesicles can be formed and targeted. We will focus primarily on the first four stages of this process and only mention system reset briefly at the end of this topic.
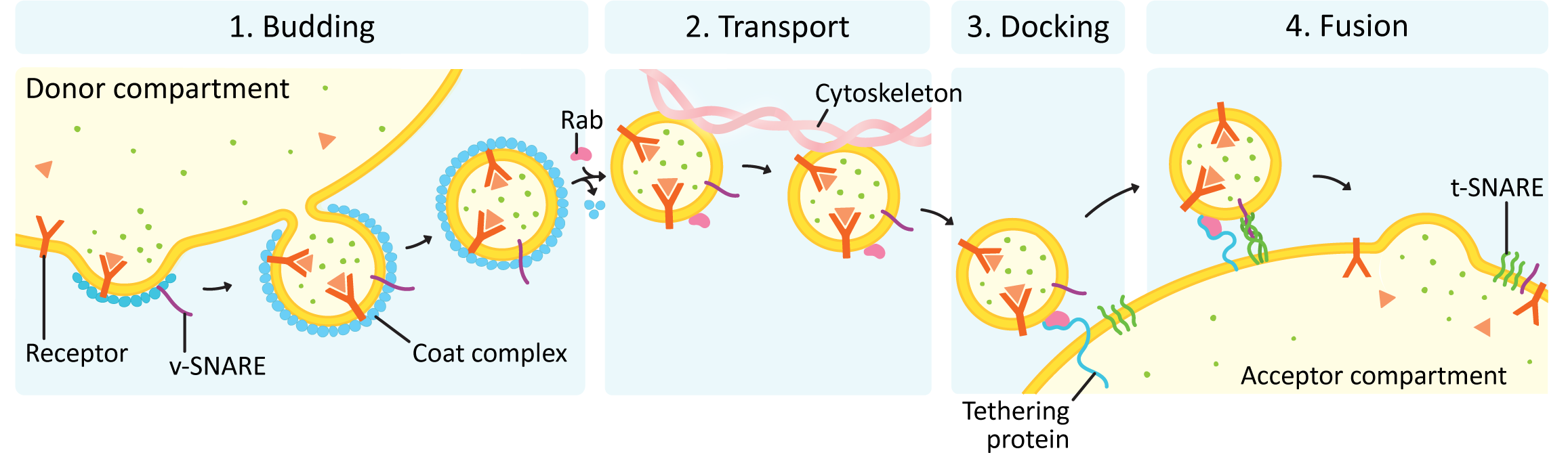
Each of these stages must be carried out correctly. There is no margin for error in vesicle traffic due to the potentially destructive nature of the cargo that is being transported. Remember that proper function of the endomembrane system is absolutely essential to survival, which means that errors may very well kill the cell. In this material, we will focus on budding, docking, and fusion. Transport of vesicles is carried out using a combination of the cytoskeleton (which will be discussed in Chapter 6) and sometimes short-range diffusion (which we are not going to discuss further).
Step 1. Vesicle Formation and Cargo Selection (a.k.a. Budding)
It is important to understand that membranes do not curve on their own. Energy input is required to change the shape of a membrane. As such, it is thought that virtually all vesicles require some kind of protein machinery to help them form. However, the exact protein machinery used in the formation of many types of vesicles has yet to be discovered. We still have a great deal to learn about vesicle traffic despite over 100 years of research on the endomembrane system.
While there is a variety of protein machinery used to help promote the formation of vesicles, we can identify specific elements that are very common when looking at different ways that vesicles can form (Figure 04-17). All of the vesicles that we are going to study require the following:
- Cargo receptor: Proteins destined for particular target organelles must be collected together in one spot so that a vesicle can be formed around them. This is accomplished by binding of the proteins to receptors in the membrane that recognize them. This helps create the specificity required for protein transport. From our online order analogy, the cargo receptor is what picks up the merchandise and puts it in the correct box and also helps ensure that no other merchandise ends up in the box by accident.
- Adaptor protein: The adaptor protein acts as a bridge between the cargo receptor and the coat protein. Usually there are more adaptors than coat proteins, so this allows for another level of specificity. Different adaptors can bind to different receptors but still form a vesicle using the same coat proteins.
- Coat protein: The coat protein provides the structure to support the bending membrane while the vesicle is being formed. There are many components in a vesicle that help determine the direction of transport, but specific coats are generally associated with particular pathways and directions of transport. We’ll see examples of this as we move through this topic.
- GTPase: A GTPase is a protein that binds GTP and hydrolyzes it. The energy released when the GTP is hydrolyzed can be used at any part of budding, though it appears to be primarily associated with the release of the vesicle coat after its job is done.
It’s also important to know that the lipid composition at the site of vesicle budding is just as important as the protein machinery. Membrane lipids found in ER exit sites, and other regions from which vesicles bud, are important in helping to assemble protein machinery, allowing the membranes to curve, and even as identity markers. So even though we will not discuss the lipids much in this chapter, you should know that the lipids are equally important to proper formation and function of vesicles.
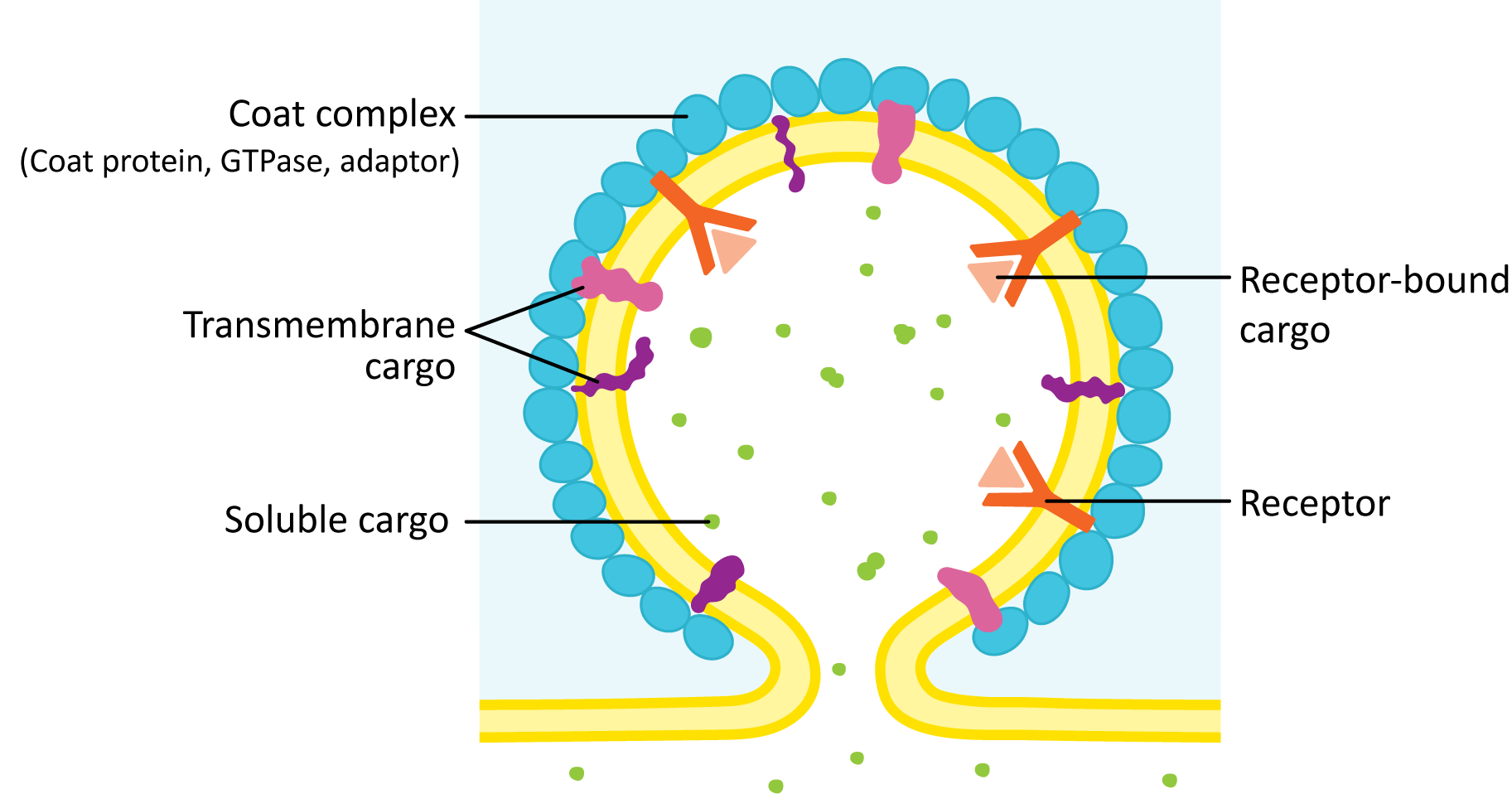
Steps in the Formation of a Vesicle
- Cargo proteins are held in place by transmembrane cargo receptor proteins.
- An adaptor protein is a protein that binds to the cytosolic side of the receptor as well as a cargo receptor protein.
- A small GTP-binding protein (called a GTPase) acts as a regulatory unit to determine exactly when the adaptor and coat proteins are allowed to bind to the receptor. It is in its active form when bound to GTP. Over time, the GTP gets hydrolyzed to GDP, which inactivates it, and the GTPase falls off the membrane.
- Proteins involved in flipping lipids across the membrane (related to the flippases and scramblases of Chapter 2) initiate the curvature, which is the start of a vesicle bud.
- Coat proteins assemble into a lattice or network on the cytosolic side of the forming bud to help stabilize and extend this curvature (budding) until a vesicle is formed.
- To release the vesicle, the forming vesicle bud needs to be cut off from the donor membrane (a process known as scission). This action may be part of the function of the coat protein, or it may be done by a separate protein.
Once the vesicle has budded off, the coat is usually removed, the vesicle can be transported to the target organelle, and the naked vesicle fuses with its target membrane.
An Introduction to the Vesicle Coat Proteins
All coat proteins have two primary functions:
- To stabilize membrane curvature in the forming bud
- To capture cargo molecules for transport
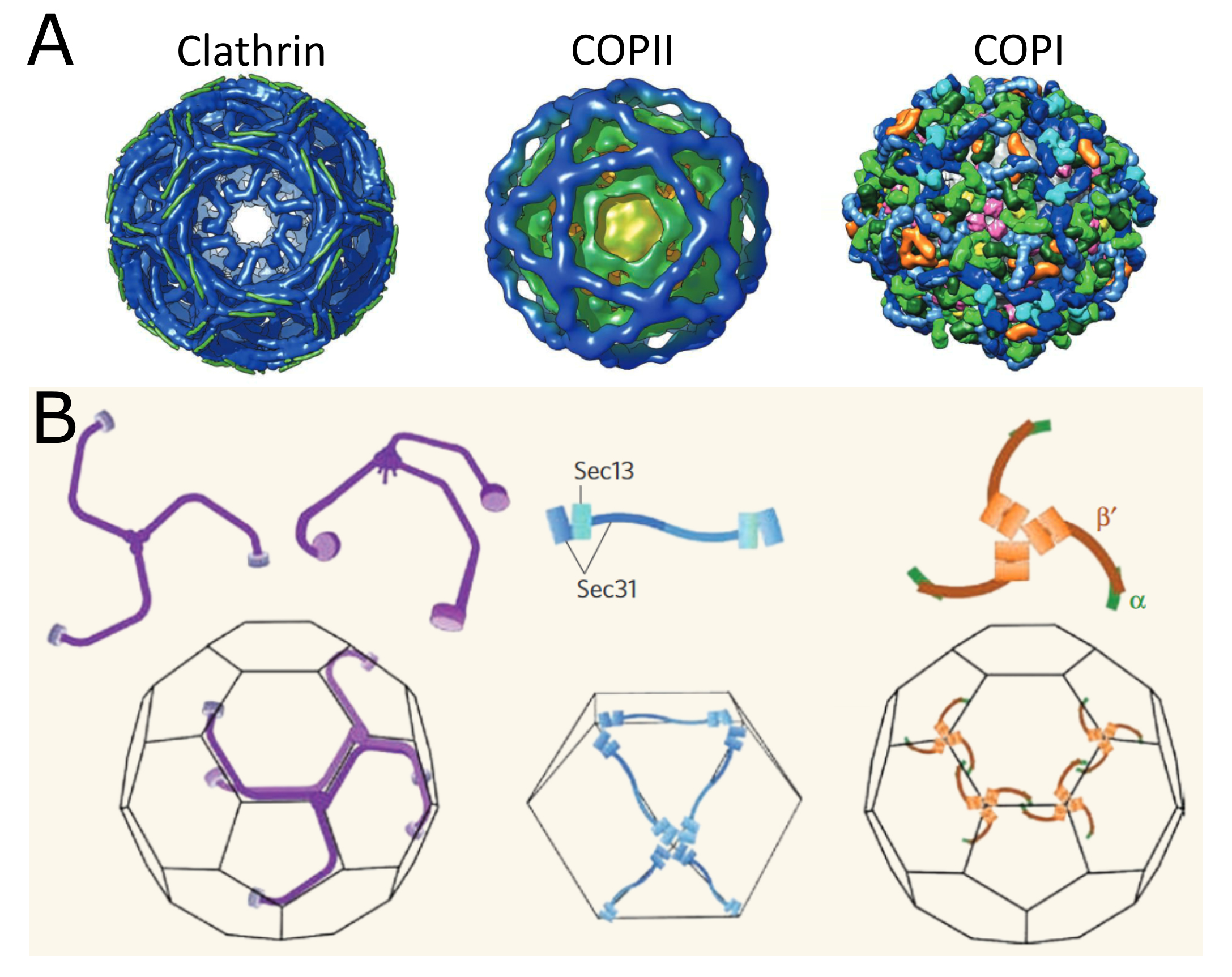
There are at least six types of vesicles where the vesicle-forming machinery is currently known and many more types of vesicles where we are still unsure how they are formed. Of the six known types, this textbook explores three of them in more detail, as they bear the most similarities to each other and are the most well characterized. More specifically, we will be looking at the three major classes of coat proteins and their role in vesicle formation. The three types of coat proteins we will study are COPI, COPII, and clathrin. Each of these types of coats tends to be used in different locations in the endomembrane system and/or for different purposes. They are also considered to be the “canonical coats,” probably because they were discovered first but also because they appear to be the most widely used by the cell.
- Clathrin coats were the first coat proteins to be discovered and as such are the ones that we know the most about. They are most commonly used between the trans Golgi network (TGN) and endosomes, lysosomes, and the plasma membrane. However, even in these sites, they aren’t always used. Clathrin coats have been shown to be important in specific kinds of traffic:
- carrying newly synthesized lysosomal enzymes from the TGN,
- bringing in molecules from the extracellular space via receptor-mediated endocytosis (and possibly some other endocytosis as well),
- recycling unnecessary membranes from secretory vesicles as they mature, and
- sending proteins out of the cell using the secretory pathway in highly regulated circumstances (like in neurons, where vesicles full of neurotransmitters are sent down to the axon terminus).
- COPI coats tend to be used for retrograde traffic, which means traffic that goes “backward” through the endomembrane system. Specifically, COPI coats are used for either Golgi-to-ER traffic or moving cargo backward through the cisternae of the Golgi. Some evidence suggests they can also be in other areas, but only in very specific situations. COP stands for “coatamer protein.”
- COPII coats are most commonly used for anterograde traffic, which means forward traffic. Specifically, they’re used for ER-to-Golgi traffic. They also have the ability to make vesicles of different sizes and shapes, unlike some of the other coat proteins, so they are used for the secretion of large molecules (like collagen, for example). As a result, COPII can also be found at the TGN, but they are not as common as clathrin. COPI and COPII coats were named in the order in which they were discovered.
In a nutshell, clathrin is primarily used in vesicles originating from the TGN and from the cell surface. COPI and COPII are mostly used as the coat proteins for vesicles originating from the ER and within the Golgi. Table 04-01 summarizes some of the structural details of these three coats as they relate to the coat components described in Figure 04-17.
| Comparing Vesicle Coatings | ||||
|---|---|---|---|---|
| Coat | Coat components | Adaptor | GTPase | Scission protein |
| Clathrin | Clathrin triskelion complex | Adaptins (AP1, AP2, or AP3) or GGA complexes |
Arf (for GGAs only) |
Dynamin |
| COPI | 7 subunits in complex |
← Included in coat | Arf | Included in coat |
| COPII | Sec13/31 complex | Sec23/24 complex | Sar1 | Included in coat |
Clathrin-Coated Vesicles: A Case Study
Since clathrin is the most extensively studied of the vesicle coats, we will use it as a case study to look at the finer details of how vesicle coats form. The clathrin cage is made from two different kinds of proteins known as the heavy and light chains. These chains come together in a specific arrangement to form a triskelion, which can be seen in Figure 04-18.
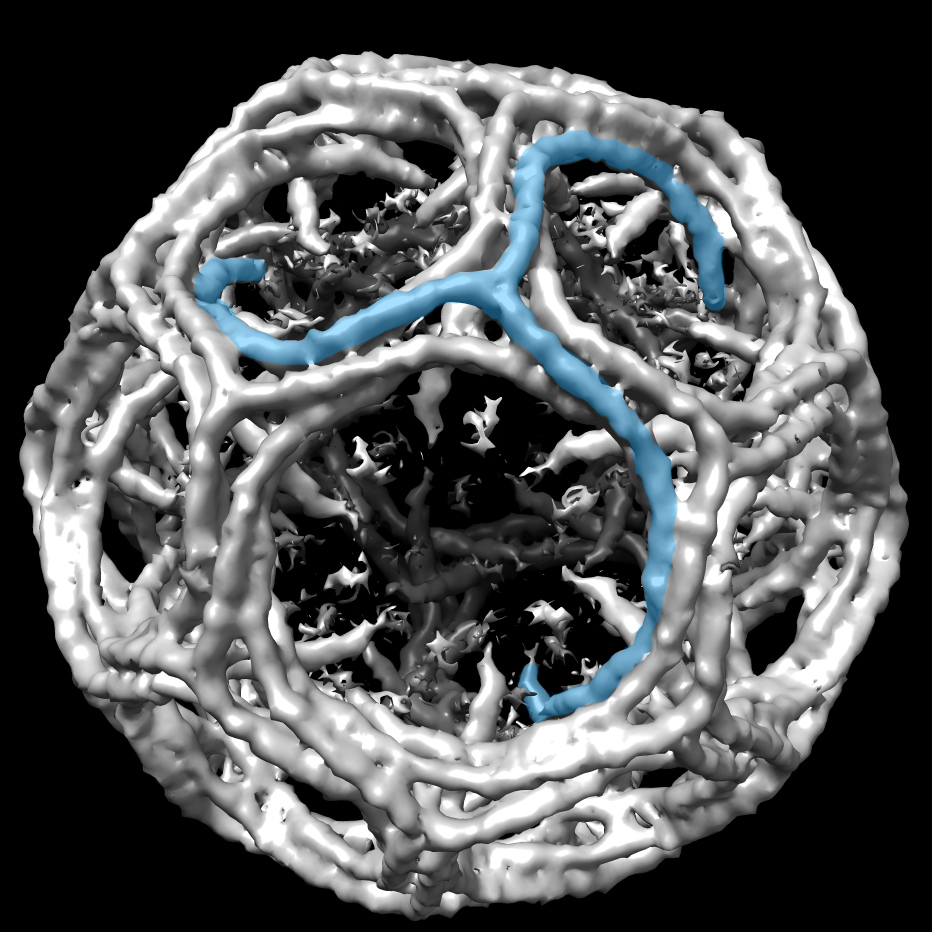
The triskelions bind to the adaptor (usually adaptin for clathrin-coated vesicles) and assemble to form the coat. As they assemble, they pull the membrane with them to form a ball. While the lattice helps shape the vesicle, it cannot cut the vesicle off from the donor membrane. A separate scission protein called dynamin is used for this purpose. Dynamin has a spiral shape and wraps around the stalk formed by the budding vesicle. Dynamin binds GTP and hydrolyzes it, which allows the protein to constrict around the membrane stalk until the membrane splits and the vesicle is released.
Clathrin works primarily to mediate traffic between the TGN, the endosomes, lysosome, and the plasma membrane. It’s important to note that not all traffic in this area uses clathrin, but most of the traffic that clathrin mediates is somewhere in this region. For example, clathrin is used in certain kinds of endocytosis, which means that the cargo is coming in from the cell exterior. It’s also used to mediate the movement of newly synthesized lysosomal proteins as they move from the TGN to the endosome, which will eventually mature into the lysosome. A notable exception is that clathrin is not usually involved in protein secretion (i.e., the default pathway out of the cell).
COPI and COPII Coats
The process of vesicle budding is essentially the same in COP-coated vesicles as it was in the clathrin-coated vesicles we just saw. In fact, all three follow the same general trajectory that we saw at the start of this section. Video 04-05 (shown below) is an excellent molecular animation of COPII vesicle formation. The different structures of the coats make for differences in the geometry of the cage that is formed and the size of the vesicle (Figure 04-19). Additionally, COPI and COPII coats both are able to promote scission themselves. As such, no additional scission protein (like dynamin) is required. Instead, a small GTPase is used as part of the coat and helps the coat release when its job is done. For COPI, the GTPase used is known as Arf, and for COPII, it’s called Sar1.
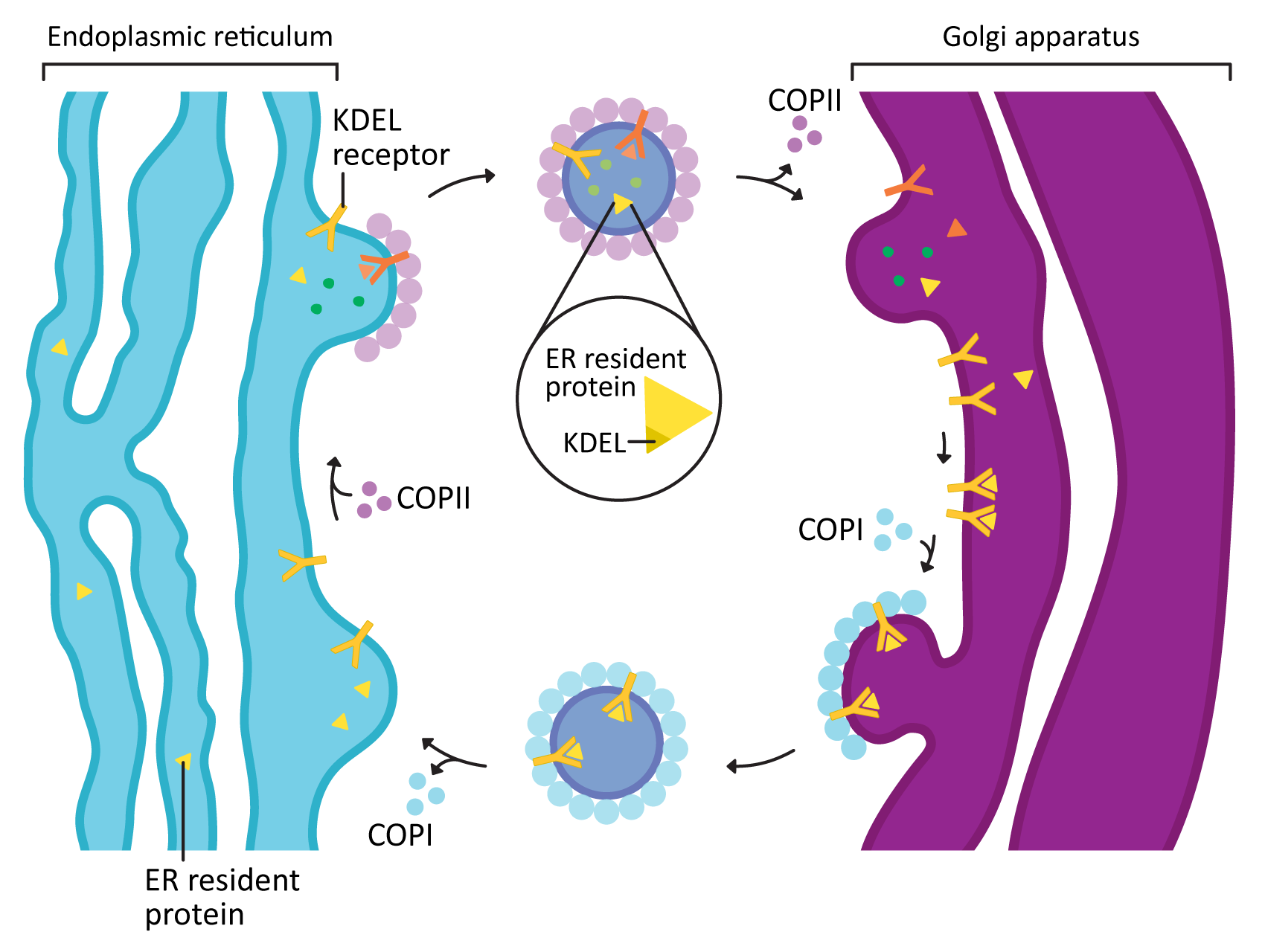
COPI and COPII mostly mediate traffic between the ER and Golgi and travel in opposite directions (Figure 04-20). This was mentioned briefly above, but this list gives you more detail:
- COPII vesicles move in the forward direction from the ER to the Golgi (i.e., anterograde traffic). As such, it usually is transporting newly synthesized proteins that need to leave the ER on their journey through the endomembrane system.
- COPI vesicles, on the other hand, mediate retrograde traffic back to the ER from the Golgi. This will include proteins that need to be returned to the ER and that are residents of that compartment. This is where the ER retention signal (also known as KDEL, first discussed in Topic 4.1) becomes important.
- The KDEL is recognized by the KDEL receptor, a transmembrane protein that lives in the Golgi where vesicles from the ER fuse.
- The KDEL receptor binds to the KDEL sequence, which is a part of the ER resident protein primary sequence.
- When the KDEL receptor binds to its cargo, a conformation change happens that exposes a COPI binding site on the cytosolic side of the protein.
- Once the COPI binding site is exposed, the COPI coat can assemble so that the KDEL receptor, with its cargo, can be packaged into vesicles and sent back to the ER.
- Once its cargo is released, the empty KDEL receptor is returned to the Golgi via COPII-coated vesicles.
Interestingly, COPI and COPII coats both have a unique additional feature, which we really only discovered in the last 10–20 years. The COPI and COPII cages are somewhat flexible in how they are arranged on the membrane. As such, they can produce larger “vesicles” that can accommodate cargo that would not otherwise fit. For example, collagen is one of the most abundant proteins in the extracellular matrix of animals, which we were introduced to briefly at the end of Chapter 2 (see Figure 02-20). Collagen is synthesized by the cell in the same way that other secreted proteins are: it is co-translationally inserted into the ER lumen, travels through the endomembrane system, and then is packaged into vesicles at the TGN to be sent to the plasma membrane for secretion. Collagen is a very large, fibrous protein, so it doesn’t fit into traditional COPII vesicles. However, COPII is able to rearrange itself into a giant tubule that is large enough to accommodate collagen so that it can be sent out to the extracellular matrix.
Other “Coat” Proteins
As mentioned earlier, our focus in this textbook is on the more traditional vesicle coats that do the bulk of the work in the endomembrane system. However, there are a number of other vesicle “coats” that are involved in the formation of vesicles that are often ignored in introductory cell biology texts such as these. We will list them here, and what they do, so that you have a more complete perspective of vesicle traffic. Also keep in mind that this list might not be complete. Like so many aspects of cell biology, this is an area of active research, and there’s always more to know!
- Caveolin helps create vesicles during endocytosis. Whereas clathrin-coated vesicles are usually used for receptor-mediated endocytosis, caveolin is used for a type of endocytosis known as pinocytosis. We’ll learn more about endocytosis (but not caveolin) in the next topic in this chapter.
- The SNX/retromer complex is primarily involved in retrograde traffic from the endosome to the TGN. In general, it forms large tubules that help in cargo selection. Then the vesicles are budded off the end of the tubule.
- ESCRT is pretty cool, as it is used to bud vesicles in a way that is opposite from most other coats. Instead of budding vesicles into the cytosol, they bud them away from the cytosolic compartment. This is usually used to push vesicles into the endosome so that membrane proteins can be fully degraded by the lysosome. Unfortunately, ESCRT is a common target for hijacking by membrane-bound viruses to help them bud out of cells so that they can go move to a new cell and restart the infection cycle. Both HIV-1 and the Ebola virus are known to do this.
While their mechanism of function is different from the more canonical coats, a few truths still hold. These protein complexes help bend the membrane and pinch off the new vesicle. The retromer and ESCRT also have mechanisms to choose cargo, whereas caveolin is considered nonspecific.
Step 2. Transport of Vesicles from the Donor Compartment to the Target Compartment
There are two possible modes of transport for a vesicle:
- Over short distances, vesicles can move by diffusion. This is thought to be quite common in mitosis of plant cells, especially when a new cell wall needs to be rapidly secreted between the two new cells. It is also known as bulk flow.
- Over longer distances, vesicles move along cytoskeletal tracks (usually microtubules but also actin) and are moved by motor proteins (kinesins or dynein for microtubules, myosin for actin).
As mentioned before, we will look at how the cytoskeleton works in Chapter 6, so we direct you there to understand how this process might work.
Steps 3 and 4. Targeting of Vesicles (a.k.a. Docking and Fusion)
Like vesicle budding, docking and fusion must be specific. Errors cannot happen or the cell might die. As such, there is additional machinery involved in making sure that docking and fusion happen accurately and efficiently at each of the different target membranes.
3. Vesicle Docking
Vesicle docking is a way of bringing a vesicle in close to the target membrane to place it in the perfect position for the final fusion step (see Figure 04-21). There are two main categories of proteins involved in this process: Rabs and tethers.
- Rabs are small GTPases that sit on the vesicle surface when activated and help identify the vesicle as one that is headed to a particular target membrane.
- Rabs are said to be used for “membrane identity.” This means that each organelle in the endomembrane system has its own sets of Rabs. If a vesicle buds from the ER, for example, any ER Rabs will be lost, and a vesicle Rab will dock. Once the vesicle fuses with the target compartment, the vesicle Rab will fall off, and a new Rab will take its place.
- Rabs are part of a larger “superfamily” of small GTPases whose members you will meet over and over again in cell biology. Ran, which you may remember from nuclear import in Chapter 3, is also part of this family, as well as Arf1 and Sar1, which help with vesicle coat assembly and disassembly in COPI and COPII, respectively. Even tubulin (used to make microtubules) is a distant relative.
- Tethers are a much more diverse group of proteins, usually with very little genetic similarity between them. However, their role is the same in all cases: to bind to the vesicle Rab and help capture the vesicle from the cytosol and pull it in closer to the membrane of the target compartment.
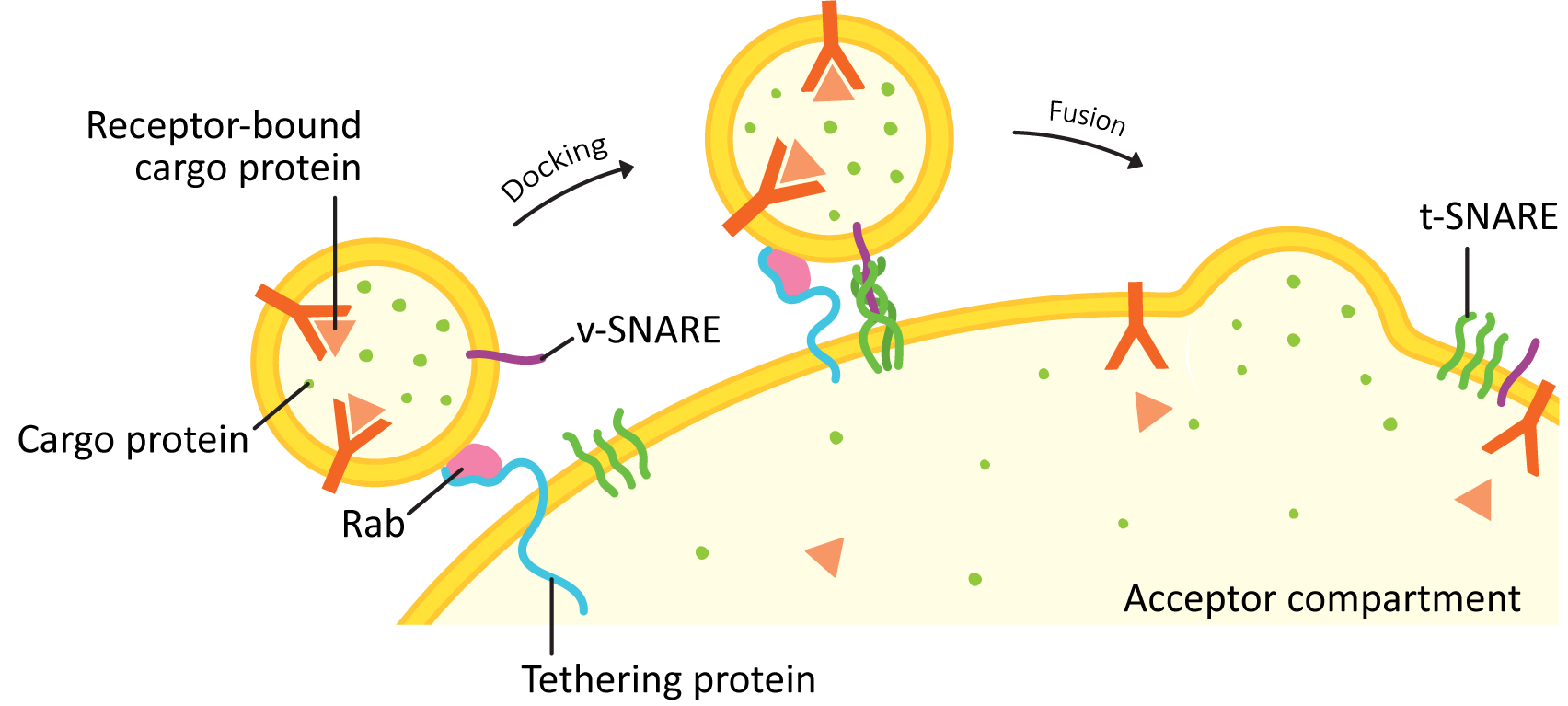
4. Fusion with the Target Compartment
Once the Rabs and tethers have done their work, the final step of this process is mediated by a family of proteins known as SNAREs. The word SNARE stands for SNAP receptor (SNAP itself is an acronym that stands for synaptosomal-associated protein). For this reason, we always write the name of the protein in all capital letters as SNARE.
SNAREs are generally categorized into two major groups: vesicle-SNAREs (v-SNAREs) and target-SNAREs (t-SNAREs). This grouping is based on their location in the cell. The v-SNARE is embedded in the vesicle membrane, and the t-SNARE is embedded in the target membrane.
Structurally, SNAREs fall into two major categories: Q- and R-SNAREs. In order for membrane fusion to occur via SNAREs, one R-SNARE and 3 Q-SNAREs must be present. Some act as v-SNAREs, whereas others will be t-SNAREs.
Once the vesicle has been tethered, the four SNARE coils interact and “zipper” together (Figures 04-21 and 04-22). This pulls the vesicle in tightly enough that all of the water molecules get pushed out of the way, and the two membranes can interact directly. Once that happens, the membrane lipids can intermingle, and the membrane will fuse. Both v-SNAREs and t-SNAREs must be present for this to occur. This binding is very specific…not any old v- or t-SNARE will do.
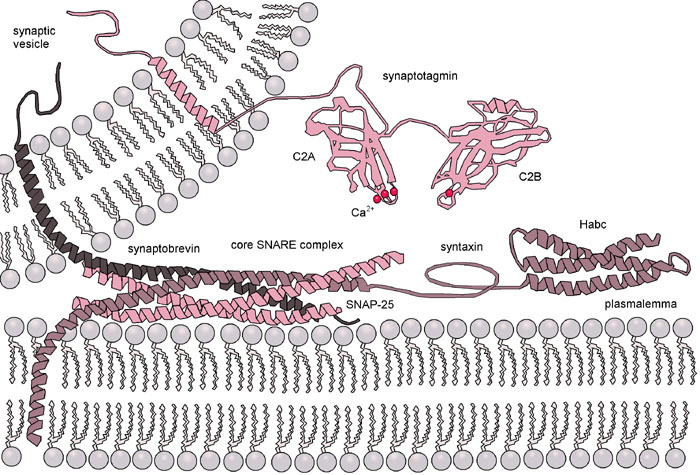
A further illustration of vesicle fusion can be seen in the Video 04-06. At the end of the video, a protein comes in and uncoils the four SNAREs from each other. This is known as resetting the system, which was mentioned way back at the start of this topic. Separating the SNAREs after fusion is vital so that they are available for the next vesicle that docks.
In addition to uncoiling the SNAREs, resetting the system involves transporting the v-SNAREs back to their original target compartment so that they can be used in another round of vesicle fusion. Transporting SNAREs back is not trivial, as you need to make sure that they don’t accidentally get used in transport. Usually, the SNAREs that are traveling as cargo get covered up by a regulatory protein (called n-Sec1) so that they can’t get in the way. But that story is mostly beyond the scope of this textbook.
Topic 4.3: The Golgi Apparatus
Learning Goals
- List the major function(s) of the Golgi.
- Explain how the structural compartmentalization of the Golgi creates different environments and how this contributes to Golgi function.
- Compare and contrast the Golgi structure and function in animals, plants, and fungi.
- Describe the sequence of events occurring during protein glycosylation in the ER and Golgi.
Introduction
As material travels through the endomembrane system, it will move from one compartment to the next, using vesicles to traffic between compartments. There is an order to the flow of traffic within this system. Newly synthesized proteins always start their journey at the ER, which was our focus at the start of this chapter. Then they are packaged into vesicles and are sent to the Golgi apparatus. From there, they may take different paths, depending on their role and destination. Some proteins will be sent to the endosome and then the lysosome, while others will head directly for the cell exterior. Even cargo that has entered the cell via the endocytic pathway (Figure 04-01) sometimes comes as far into the cell as the Golgi before being redirected to another destination.
Just like the ER, and the other organelles of the cell, the Golgi apparatus has multiple roles. It is often considered to be the heart of the endomembrane system, as it receives cargo from all directions (ER, plasma membrane, endosomes), modifies it, and then repackages it into new vesicles to be sent to new destinations. It is also the site of all of the major cellular work involving polysaccharides. While some of the work starts or ends elsewhere in the cell, the Golgi is the site of virtually all of the known glycosyltransferases (i.e., enzymes capable of covalently attaching sugars to each other). Not only is this important for the production of glycoproteins (i.e., proteins with sugars attached), but it is vital for the production of the cell walls in plants, algae, and fungi. In this topic, we will specifically focus on the structure and function of the Golgi.
Structure of the Golgi
The Golgi was named after an Italian researcher by the name of Camillo Golgi in the late 1800s. He discovered the Golgi during his exploration of the central nervous system by microscopy. (It is for this reason that we always write the name of the organelle with a capital G.)
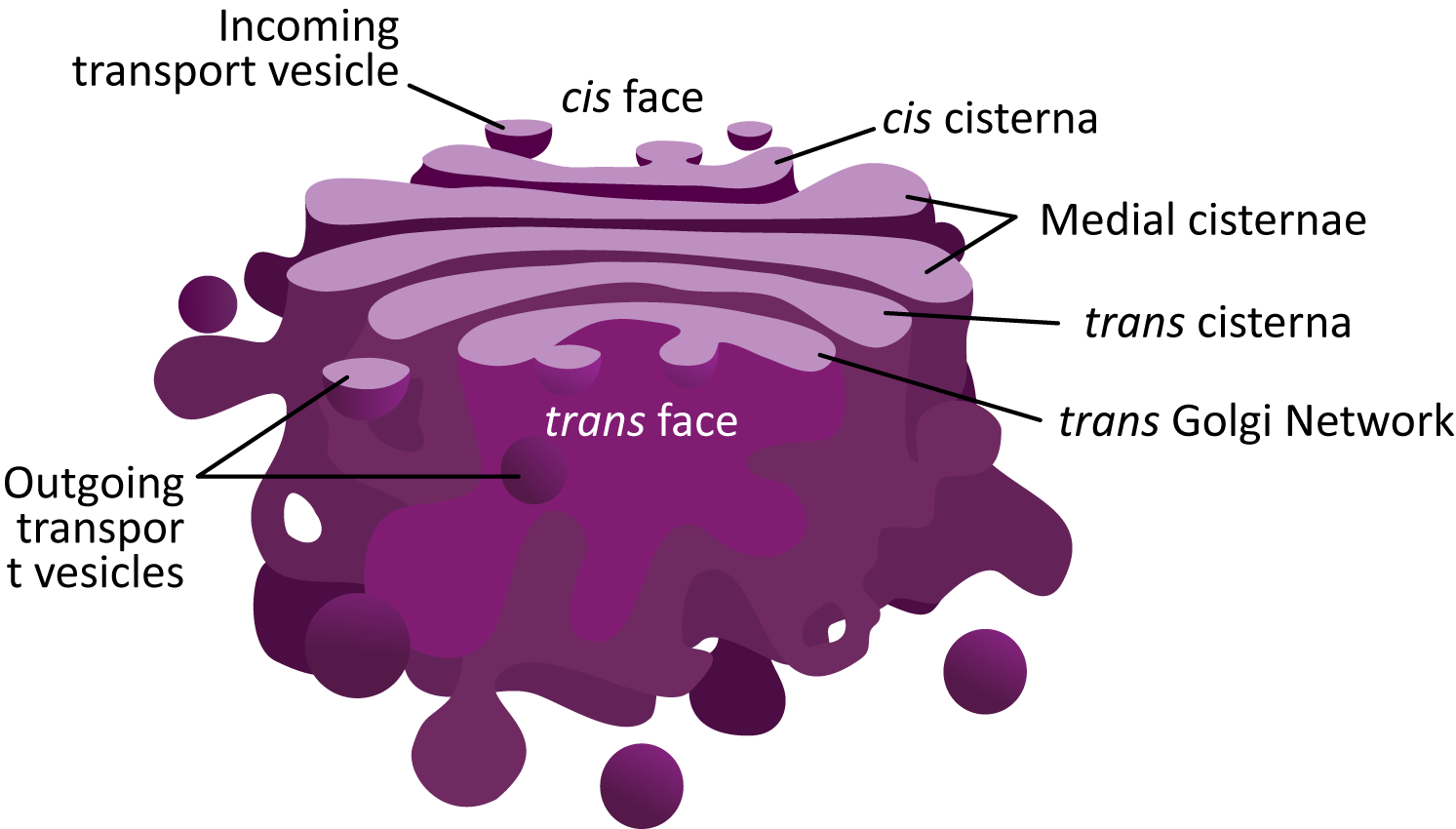
Figure 04-23 showcases the common structural features of the Golgi and highlights many of the new terms you will need to know to discuss this organelle. The following are some important points to note:
- Each of the “pancakes” of the Golgi is called a cisterna (plural cisternae).
- The interior space of each cisterna is known as the lumen. You have seen this term before, as it is also used to identify the space inside the ER, called the ER lumen.
- The Golgi is considered to be a “polar organelle” in that cargo enters at one end (the cis face, for newly synthesized proteins) and exits at the other end (the trans face). As such, we identify each of the cisternae by their location within the stack:
- The cis cisternae are the ones nearest the cis face, which receive vesicles from the ER.
- The medial cisternae are in the middle of the stack. There can be more than one.
- The trans cisterna is closer to the trans face of the Golgi stack.
- The trans Golgi network (TGN) is the trans most cisterna of the sac. Often it is more convoluted, as vesicles will be budding and fusing at this site. We’ll hear more about the TGN later.
The structure of the Golgi apparatus is both surprisingly variable between different kingdoms/species and yet also very similar. For example, in all eukaryotes, the Golgi is made up of a series of flattened sacs that work together like an assembly line, moving cargo through them in order. However, the location and arrangement of those sacs can differ.
- In animals, the cisternae of the Golgi are large and are located centrally, near the nucleus (thus we say they have a perinuclear position). Usually, there is only one, or maybe two, Golgi in an animal cell.
- In plants, on the other hand, Golgi stacks are very small and tumble through the cytoplasm, using a combination of cytoplasmic currents and the actin cytoskeleton to help them move through the cell. There can be hundreds of these tiny Golgi stacks in a plant cell.
- In yeast and other fungi, The Golgi arrangement is quite variable. In some fungi, it is more “animallike,” while in others, it is more “plantlike.” In still others, it is different yet again. In the yeast Saccharomyces cerevisiae, the individual cisternae of the Golgi move around the cell independently of each other.
In this chapter, we will primarily focus on the features of the Golgi that are consistent across kingdoms, with only a few highlights of the unique characteristics specific to particular cell lineages.
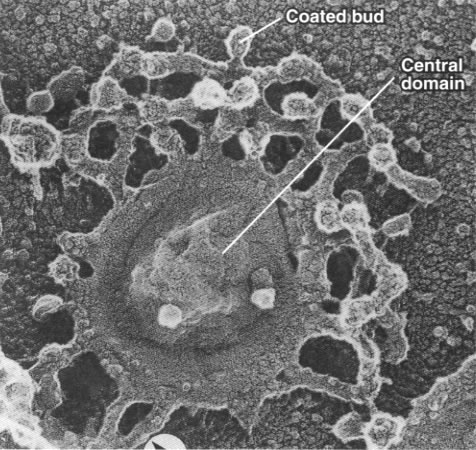
While the cisternae do look like flattened sacs in cross section (Figure 04-23), there is more complexity to the structure. Vesicles are regularly budding and fusing with the margins of the cisternae as material is moved forward or backward within the Golgi stack. As such, if you were to look at a single Golgi cisterna from the top, it would look a bit more like a “paint splat” than a nice round pancake. This can be observed in the electron micrograph below (Figure 04-24).
Localization of Resident Proteins Can Be Quite Specific
Within the Golgi there is a net flow of newly synthesized proteins (i.e., the cargo) from cis to trans. In addition, each part of the organelle has specific proteins that are residents in that region. For example, there are specific proteins that are located in the cis cisterna of the Golgi that are required to remain there in order to carry out their function. The same is true for the enzymes that are found in the medial cisternae, the trans cisterna, and also for those that locate to the TGN. This arrangement can be visualized exceptionally well using immunolabeling and a technique known as electron tomography (Figure 04-25). Tomography is a specialized technique that allows us to create 3D models from the sample sections we normally use in an electron microscope. In each of the panels of Figure 04-25, we see an example of a 3D reconstruction of a Golgi stack with different resident enzymes that have been labeled using gold particles attached to an antibody. The colored dots (yellow in A and C; red in B) represent that gold particle label.
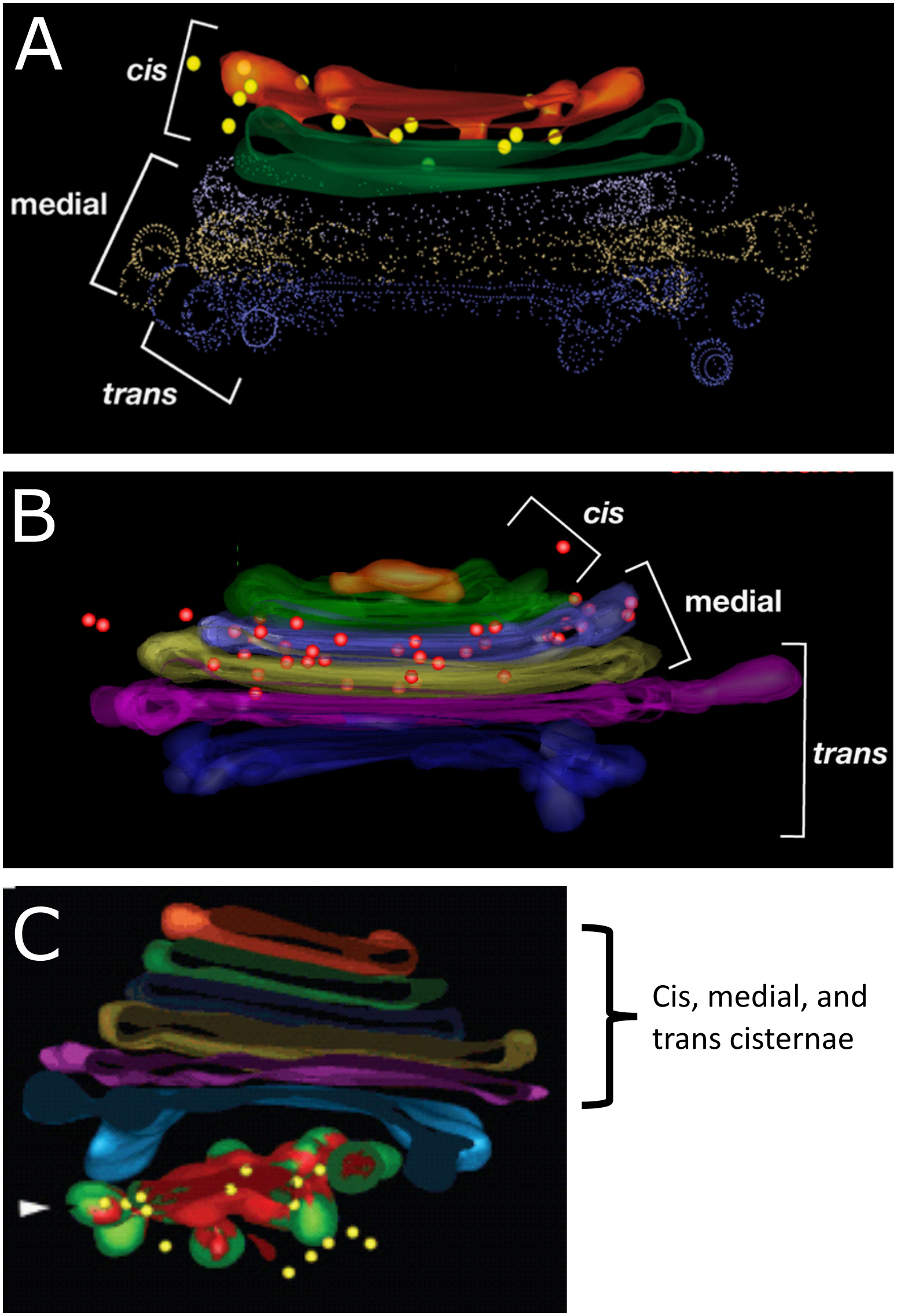
Just like we’ve seen with other organelles, the ability of the proteins of the Golgi to localize so specifically can be traced back to the order of amino acids found within their primary sequences. There are specific sequences that are used to allow proteins to become part of a COPI- or COPII-coated vesicle and other sequences that allow proteins to be retained as a resident in the cis, medial, or trans Golgi (instead of moving through the cisternae and out the other side). As we learn more about the Golgi, we find that the specific localization of individual resident proteins might be even more precise, not only living within a single cisterna but also located in a specific region of that cisterna (inner versus outer ring). While you are not expected to know the details about the specific composition of these targeting sequences, it is important that you recognize that the ability of these proteins to localize so precisely is based on their amino acid sequence and can be traced right back to the DNA code they were made from.
Golgi Dynamics: How to Get from One Cisterna to the Next
One of the biggest challenges that the Golgi faces is that some proteins are “transient” and must move through the Golgi (i.e., from cis to trans), whereas other proteins are “resident” and need to stay where they are inside a given cisterna. As If that weren’t challenging enough, vesicles constantly fuse and bud from the different cisternae, adding and removing proteins and lipids from the membrane. So how does the Golgi maintain its resident proteins in the proper location while allowing other cargo to be pushed along through the system? Scientists have been trying to solve this dilemma for almost as long as the detailed structure of the Golgi has been known.
Over the years, a number of ideas have been put forth to explain this. As we learn more, different ideas have gained and lost favor. Currently, the most widely discussed models for transport through the Golgi are called the cisternal maturation model and the vesicular transport model (sometimes called the stable cisternae model). See Video 04-07 for an excellent explanation of the two.
While the two models are presented as separate possibilities, the current thinking is that the method by which proteins are moved and processed in the Golgi is probably more complex than any one model predicts (not really a surprise). The most widely accepted model right now is the cisternal maturation model (Figure 04-26), even though there is clear evidence that at least some things move via vesicular transport. The cisternal maturation model assumes that new cis cisternae are continually formed by the fusion of vesicles that are flowing to the cis Golgi network from the ER. The previous cis most cisterna is now a medial cisterna, as it has been pushed further along in the stack. In this model the trans cisterna is the oldest and it started life as a cis cisterna. The trans cisterna breaks up into tubules and vesicles to form the TGN.
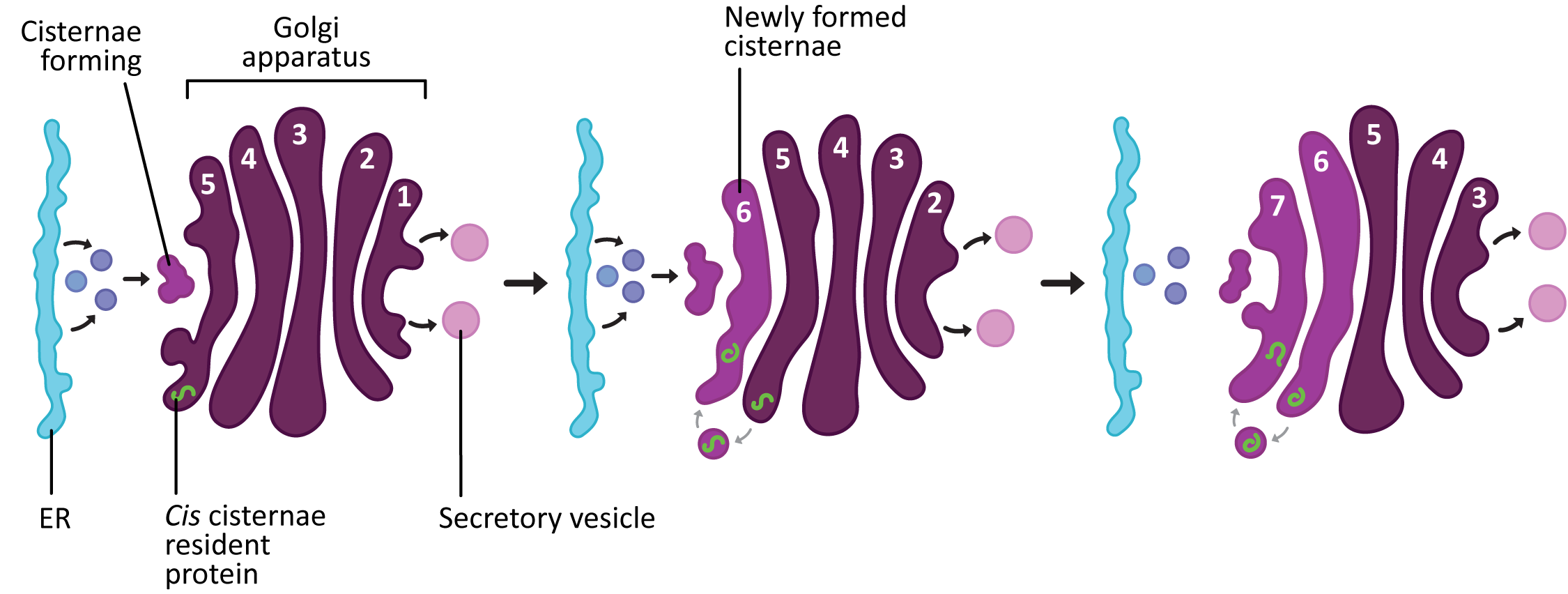
According to the cisternal maturation model, the proteins that need to pass through the Golgi (like newly synthesized proteins) stay in place inside the cisternae and simply enjoy the ride as they move from cis to trans. On the other hand, since the different cisternae also have different functions, “becoming” a medial cisterna will require that the resident proteins that work in the cis cisternae be removed from the new medial cisternae and sent backward to the newly formed cis cisternae. This retrograde trafficking is thought to be achieved via COPI vesicles.
Protein Processing in the Golgi: Glycosylation
One of the most common posttranslational modifications of proteins is the addition of polysaccharides. This process is called glycosylation (Figure 04-27). The enzymes that carry out these reactions are located in the lumen of the ER and the Golgi apparatus and not in the cytosol. This means that the proteins being glycosylated are residents of one of the organelles of the endomembrane system, are bound for secretion, or are membrane proteins.
Glycosylation serves a variety of structural and functional roles for proteins. Inside the endomembrane system, it can serve as a targeting sequence, an indicator of proper folding, and a signal for receptor binding. Cell surface proteins are almost always glycosylated, as they were secreted via the endomembrane system. The surface oligosaccharides have multiple roles, including signaling, defense, cell-cell adhesion, and more. For example, immune cells specifically recognize a special glycosyl variety called lectin to move amongst tissues during immune surveillance. In another example, viruses like Human Immunodeficiency Virus (HIV) and the COVID-19 virus (SARS-CoV-2) also use heavily glycosylated proteins as a shield against the immune system to evade detection. Thus, glycosylation is very important and serves wide ranging roles in cells.
There are two major types of glycosylation: N-linked glycosylation and O-linked glycosylation. Most introductory cell biology textbooks focus primarily on N-linked glycosylation, as it is believed to be the more common of the two. The biggest difference between the two types of glycosylation is how and where the sugar is attached to the protein. In N-linked glycosylation, the sugars are added to an asparagine (which has the 1-letter code N) within a specific amino acid sequence, whereas in O-linked glycosylation, it is a serine or threonine that gets used to covalently link the sugars (specifically, an oxygen in the R group gets used, hence the name O-linked). Otherwise, the process is somewhat similar in each case.
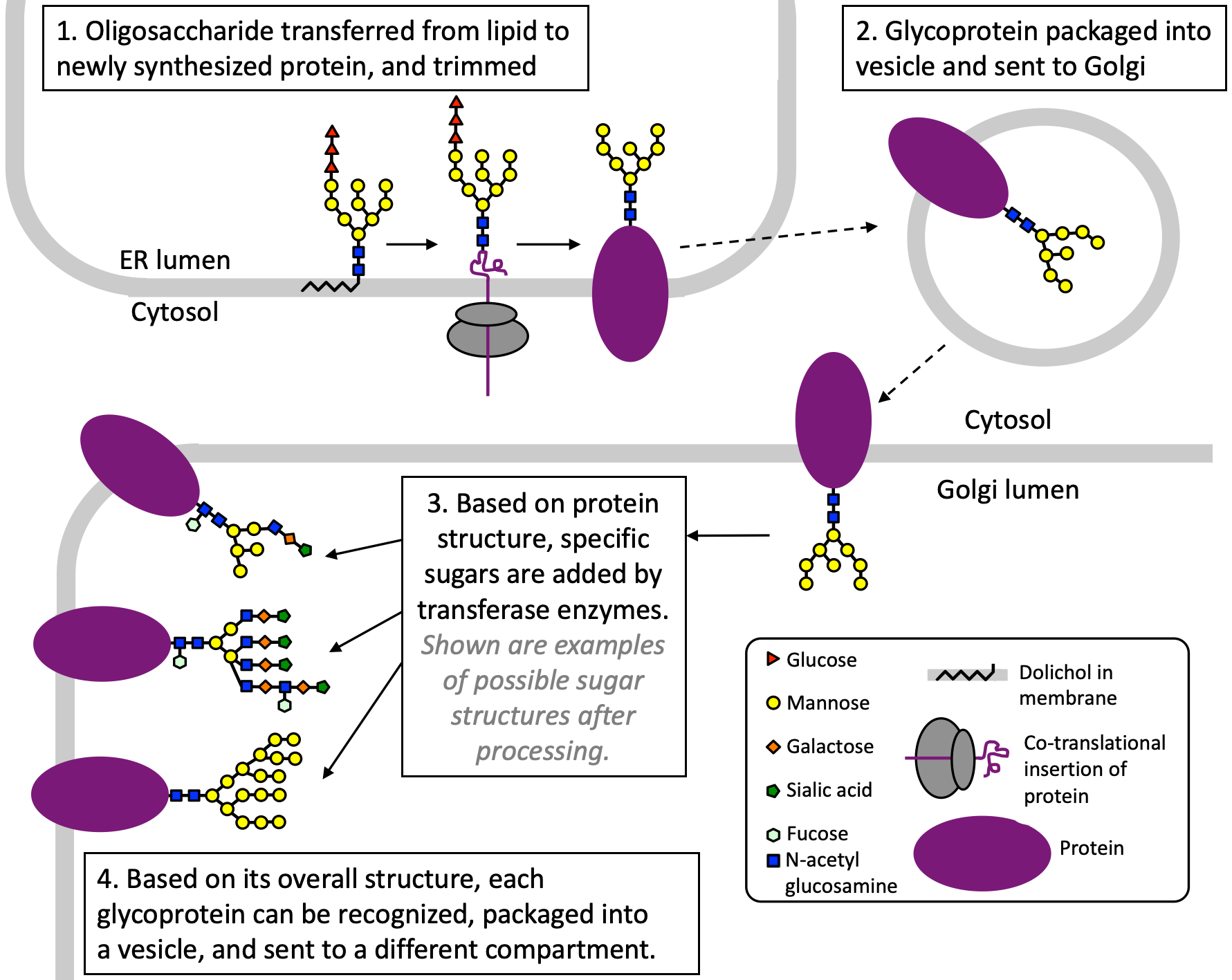
Glycosylation begins in the ER. In the initial step of the process, a prefabricated oligosaccharide “tree” consisting of N-acetyl-glucosamine, several mannose sugars, and a few glucose residues is transferred from a lipid known as dolichol (Figure 04-27, Step 1). Like all other lipids, dolichol is assembled on the cytosolic side of the ER membrane and then flipped to the lumen (cisternal) side of the membrane by a flippase (see Chapter 2, Topic 2.2). After the tree is transferred to the protein, the dolichol is free to be reused by adding a new oligosaccharide tree to it for another round of glycosylation.
After the “tree” is added to the protein, the sugars can be trimmed in the ER before being packaged into vesicles and sent to the Golgi apparatus. It is inside the Golgi where most of the glycosylation work happens. Each glycosyltransferase mediates the formation of a single type of bond between specific sugars (i.e., glucose, galactose, mannose, etc.). Thus, sugars are added one by one as the protein moves through the Golgi, like an oligosaccharide assembly line. Different glycosyltransferases reside in the different cisternae, which means that a glycosyltransferase that resides in a later compartment will be unable to add its sugar if a step in an earlier compartment does not happen. The result is an array of possible oligosaccharides that can be built onto the proteins, which can then be used for specific functions later on.
Cell Wall Production in the Golgi Apparatus
Of the various eukaryotic kingdoms, several of them include cells that are commonly surrounded by a cell wall (specifically, plants, algae, and fungi). Animals do not have cell walls, and protists are extremely variable in whether they have cell walls, not to mention what those walls are made of, which makes them a topic for a whole course on their own. The cell walls of plants, algae, and fungi are made primarily of polysaccharides. Since the Golgi is the location of virtually all of the glycosyltransferases that create new covalent bonds using sugars, it has a major role to play in the production of cell walls.
Cell walls are made of complex, often highly branched sugar chains that create a gel matrix in which cellulose is embedded. We saw examples of this before, in Chapter 2 (see Figure 02-20). These complex polysaccharides are synthesized one sugar at a time as the molecules move from cis to trans inside the Golgi, in a way similar to what we just saw for glycoproteins. They are packaged into vesicles in the TGN and then shipped to the plasma membrane, where they are secreted and then incorporated into the existing plant cell wall. The most common cell wall compound made in the Golgi is pectin, which is a highly branched molecule that helps hold all of the parts together.
The cellulose is not created in the Golgi but instead uses a special protein structure at the plasma membrane known as a rosette.
The Trans Golgi Network (TGN) Is a Major Cargo Sorting Center for Secretion
Once the proteins arrive in the TGN, it means that they have been through all of their needed processing and are ready to be sent to the next place / final destination. The primary function of the TGN is to act as a sorting center. Cargo moving through the Golgi will end up at the TGN, and based on the structure of the protein and/or the carbohydrates attached to it, it will be sorted into one of two major routes:
- The secretory pathway (a.k.a. exocytosis): This pathway is for proteins that are destined either to be excreted into the extracellular space or to become an integral membrane protein in the plasma membrane.
- The lysosomal pathway: As its name states, the lysosomal pathway is used for proteins that are residents of the lysosome.
As a result of its role, the TGN is less “pancake-like” than the other cisternae of the Golgi. In electron microscopy, vesicles are often captured in a variety of states of budding, making the TGN somewhat bulbous (Figure 04-23). There is quite a lot of activity at the TGN, as it ensures that each protein gets packaged into the correct transport vesicle so that it can be sent off to the correct destination for its function.
Interestingly, despite the fact that we spent a whole topic examining vesicle formation using coat proteins, our understanding of how cargo gets packaged into vesicles at the TGN is vague. In some cases, the structures produced are too large to be packaged into standard coated vesicles. There have been debates as to whether the TGN is able to peel off from the rest of the Golgi stack and act as its own transport compartment. In addition, recent findings show that both COPI and COPII coats are able to produce larger tubules as well as smaller vesicles, though it is still unclear how often this happens. Additionally, there are other cases where there is no evidence for any known coat protein being involved. Since the trafficking of vesicles at the TGN is an essential cellular process, it is very difficult to study. There are still a great many unanswered questions in this field.
Topic 4.4: Pathways of the Endomembrane System
Learning Goals
- Describe the secretory, lysosomal, and endocytic pathways and trace the path of cargo through each of these pathways.
- Differentiate between the functions of endosomes and lysosomes, and describe the structural relationship between the two compartments.
- Compare and contrast between constitutive and regulated versions of secretion and endocytosis.
- Interpret the results of the different experimental tools studying traffic through the endomembrane system.
After newly synthesized proteins have traveled from the ER to the Golgi, they hit a crossroads at the trans Golgi network (TGN), where their paths diverge, depending on destination and function. As such, this is a good time to step back and take a look at the bigger picture of the endomembrane system and how proteins and other cargo travel through it. Thus, in this last topic of the chapter, we will discuss each of the major pathways that cargo takes through the endomembrane system. As is our practice in this textbook, we will end by exploring the experimental techniques scientists use to study how cargo moves through the different pathways.
First, to remind you, we will be looking at three primary pathways through the endomembrane system. They were originally identified way back in Figure 04-01:
- The secretory pathway, which is also often called the “default pathway” through the organelles. It is the path that most newly synthesized ER-targeted proteins will take through the endomembrane system on their way out of the cell.
- The lysosomal pathway, which is the path that newly synthesized digestive enzymes will take to get to the lysosome, which is their final destination.
- The endocytic pathway, which is the path inward from the cell exterior for proteins and molecules brought in by the cell. Material brought in via endocytosis will also likely end up in the lysosome, where the macromolecules are broken up into their basic building blocks to be reused in biosynthetic pathways.
The Secretory Pathway (a.k.a. Exocytosis)
As mentioned above, proteins that are destined to be sent out of the cell (or to live in the plasma membrane) have two different versions of the secretory pathway that they might take (Figure 04-28):
- In regulated secretion, proteins are packaged into vesicles that are stored in the cell until they are secreted in response to a specific signal.
- In constitutive secretion, vesicles continuously form and carry proteins from the Golgi to the cell surface. This is also sometimes called the default secretory pathway, as it requires no special targeting sequence (beyond the ER insertion sequence required to first enter the endomembrane system) for cargo to get secreted via this pathway.
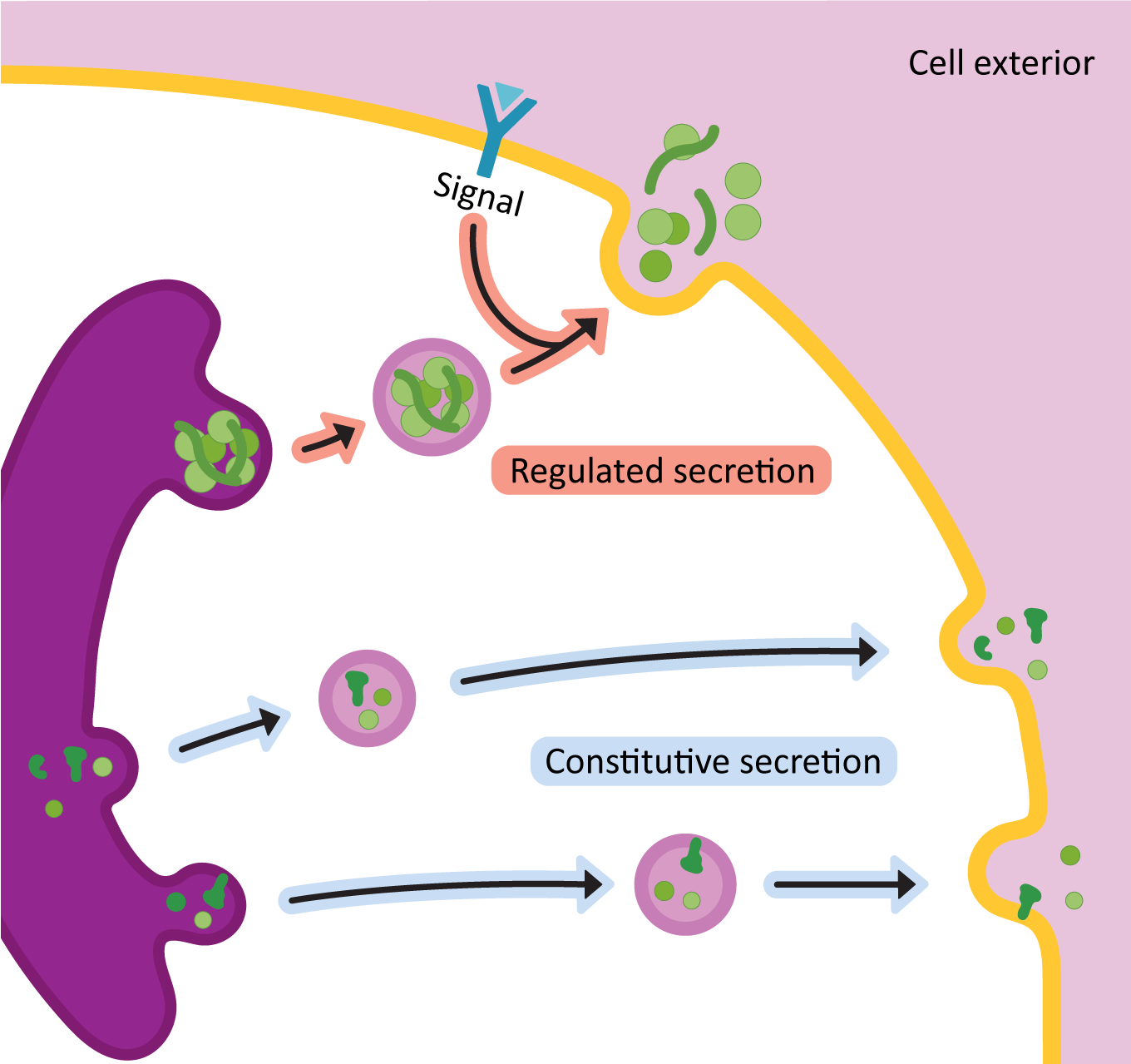
With two pathways that both lead to the plasma membrane and out of the cell, why might a particular protein be secreted via one pathway or the other? Consider, for a moment, why one protein’s exit from the cell might need to be controlled and another one might not. Can you think of examples of proteins that might be better served by traveling through one pathway or the other? These are questions to keep in mind as we explore the different forms of secretion.
Constitutive Secretion
The constitutive exocytic pathway operates continually in all cells and supplies a continuous stream of vesicles containing lipids, proteins, and polysaccharides to the plasma membrane for release outside of the cell. When we think of “secretion,” this is often the pathway we are referring to. Because it exists in all cells, and because it appears to require no specific signal for cargo to enter this pathway, we generally consider this to be the default pathway through the endomembrane system. A protein that is able to enter the endomembrane system will contain an ER insertion sequence. However, with no other targeting sequences, it will eventually get secreted out of the cell. Examples of components that are secreted via the constitutive exocytic pathway are the following:
- extracellular matrix components (in animals),
- cell wall components (in plants and fungi), and
- antibodies in activated, antibody-producing B-cells.
Cells that are currently undergoing a great deal of constitutive secretion often can be identified in transmission electron microscopy (TEM) due to the large amount of ER in the cytosol (especially rough ER).
Depending on the cargo, it is thought that constitutive secretion may, or may not, require coat proteins. Clathrin-coated vesicles have been shown to be involved in some cases, but there are other cases where clathrin is most definitely not involved. As we said, sometimes COPI or COPII gets involved, though this seems to be mostly for large cargo (such as collagen and other large extracellular matrix components) that is far too big to fit into a clathrin-coated vesicle. In some cases, we have evidence that the vesicles form and that transport is happening, but we have no understanding of how the vesicles are forming. Thus, this is another area of active and ongoing research.
Regulated Secretion
Like the name says, the release of vesicles carrying cargo in the regulated secretory pathway requires a stimulus from either inside or outside of the cell before the final fusion of vesicles with the plasma membrane is allowed to take place. The vesicles form and then sit in the cytosol until they are ready. As a result, in TEM, a high concentration of vesicles in the cytosol is considered to be a marker of a cell that is involved in regulated secretion.
It is thought that vesicle formation in regulated secretion is aided by the proteins that use this pathway for export. There is evidence that the cargo proteins tend to “clump together” in acidic conditions (we call this aggregation, and the “clumps” are known as protein aggregates). This is helpful, as the Golgi apparatus tends to become more acidic from the cis to trans sides of the stack (from a pH of about 6.8 in the cis cisternae to as low as 6.3 in the trans cisternae). The result is that in the TGN, we find these large protein aggregates, which, if large enough, can then push up against the membrane and help promote curvature. Like the constitutive pathway, none of the known vesicle budding machinery seems to be involved, so these protein aggregates are likely extremely important to the formation of vesicles in regulated secretion.
The regulated secretory pathway is used for products that must be secreted very quickly, but only on demand. For example, insulin is produced by the beta cells of the pancreas and stored in dense core secretory granules. When blood sugar increases to a threshold level, insulin-containing secretory granules fuse with the plasma membrane, releasing insulin into the blood. Some other examples of regulated secretion are the release of neurotransmitters in the nervous system and the release of breast milk during infant feeding.
In some cases, regulated secretion allows for further processing of the protein after it leaves the Golgi but before it gets sent out of the cell. For example, many of the secreted digestive enzymes in our gut are further processed after being packaged into vesicles. Often this involves a section of the protein being removed, which activates it. Our previous example, insulin, is another protein that undergoes processing while it is in vesicles, waiting to be secreted.
The Lysosomal Pathway
The other major pathway for proteins to take from the TGN is what we call the lysosomal pathway. The lysosome is a specialized compartment that is the site of intracellular digestion. Lysosomes contain a mixture of some 40 different types of digestive enzymes, including those that degrade nucleic acids, proteins, carbohydrates, and lipids.
Like the Golgi, there is quite a lot of variation in the lysosome and endosome structures in different tissues and eukaryotic species. Plants are able to use their vacuole as a lysosome instead of having a separate compartment for it. In white blood cells that engulf bacteria as part of our immune response, the lysosomal enzymes are delivered to the large phagosome directly, using vesicles, rather than via the mechanism we will describe here.
It is also worth noting that lysosomes are but one of many different membrane-bound compartments that exist in this “post-Golgi” region (meaning the various destinations that the TGN may be sending cargo to after it leaves the Golgi). Lysosomes develop from another “post-Golgi” compartment, known as the endosome, using a mechanism that is similar to the cisternal maturation model of the Golgi. There are several different endosome-like compartments that are recognized, and discussed by scientists, based on their functions. The difference between the various compartments is fuzzy at times, as one compartment can “become” another compartment through the addition and removal of specific resident proteins. For example, we sometimes differentiate endosomes as “early endosomes,” which are not as acidic, and “late endosomes,” which are more acidic and have more characteristics of the lysosome that they will eventually become.
There are also recycling endosomes, multivesicular bodies, and a compartment called the endo-lysosome. While we don’t have time to go into all of the details of each of these compartments, this makes the point that we do our best to give you a set of well-defined “rules” to understand the processes we’re exploring. However, we cannot completely remove the complexity, as it underlies everything that we are going to talk about. The function of this part of the cell is a bit fuzzy, somewhat confusing, and also very likely species specific.
Targeting Proteins to the Lysosome
Like all of the other organelles we’ve explored, in order for a protein to be directed to the lysosome, it must have a specific sorting signal within the chemical structure of the protein that the cell can recognize. Lysosomal targeting is somewhat unique compared to the other targeting signals covered so far, as it is actually a carbohydrate signal that is attached to the protein via N-linked glycosylation (described previously in Figure 04-27). The carbohydrate signal used for lysosomal targeting is called mannose-6-phosphate, or M6P. As the name implies, the M6P signal is a six-carbon mannose sugar, which has a phosphate group attached at the 6-carbon. The M6P is part of a larger oligosaccharide that was added during N-linked glycosylation and is usually at the end of the chain that is farthest away from the protein it’s attached to. These modified mannose sugars can be identified by receptors in the TGN so that they get packaged into vesicles and sent to the endosome (Figure 04-29).
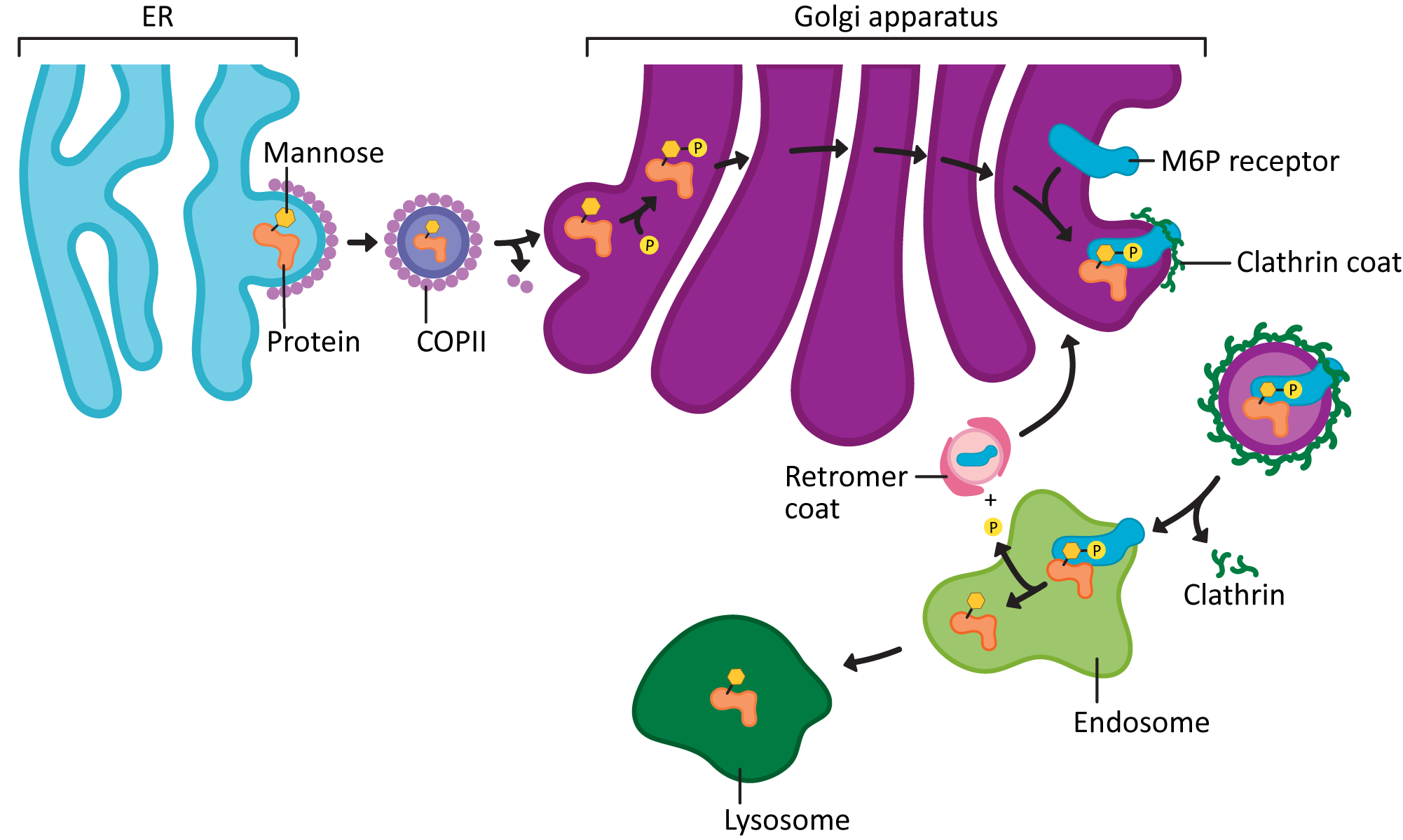
Here’s how it works:
- N-linked glycosylation occurs as usual, with the oligosaccharide “tree” being added in the ER and trimmed.
- In the cis Golgi, a phosphate is added to the sixth carbon of a specific mannose in the tree. The result of this is twofold:
- The M6P tag has been formed, which will be used later by the receptor.
- This also stops other glycosyltransferases in the Golgi from further modifying the sugar “tree.” That way the M6P tag isn’t masked or destroyed.
- In the TGN, the M6P is exposed so that it can bind to the mannose-6-phosphate receptor (M6PR).
- The protein-receptor complex gets packaged into clathrin-coated vesicles, and the vesicles are sent from the TGN to the endosome.
- There, the lysosomal proteins are separated from their receptors (due to the increased acidity in the endosome).
- The lysosomal proteins stay in the endosome, which will eventually mature into the lysosome, and the now-empty M6P receptors are recycled back to the trans Golgi network using a unique type of vesicle called the retromer.
Function of the Lysosome
The function of the lysosome is simple: to break down cellular or extracellular material into its basic building blocks. These materials could include large polymers and other components brought in from the exterior via the endocytic pathway. Lysosomes are also involved in the breakdown of bacteria that are engulfed by the cell during a process known as phagocytosis as well as in the destruction of internal components that no longer function (such as a broken-down mitochondria). As a result, in some cases, the lysosome itself is the site of digestion, while in others (such as phagocytosis), the lysosome fuses with another compartment, which releases the digestive enzymes into the new compartment to aid in the digestion of larger structures.
The lysosome is a very acidic compartment. It usually has a pH in the range of 4–5. The acidic pH of the lysosome (and the endosomes they are derived from) is maintained by membrane-bound ATP-driven pumps that move protons into the lysosomal lumen. The lysosome has a single membrane that surrounds it. Additionally, all of the proteins in the lysosome are heavily glycosylated. This makes sense for two reasons:
- First, the targeting signal is a carbohydrate signal (M6P), so the only proteins that can get targeted to the lysosome are, by definition, glycosylated proteins.
- Second, carbohydrate coverings are often used as protection in extreme environments such as this one. For example, your stomach is also an extreme, highly acidic environment that has a very similar function to the lysosome, except on a much larger scale! The cells that make up the stomach lining are protected from harm by a thick layer of mucus. Mucus is simply made up of secreted complex carbohydrates. In the lysosome, instead of secreted carbohydrates, the proteins themselves are heavily glycosylated, which provides protection to the lysosomal membrane from the digestive enzymes within it.
One more thing to reiterate: in many organisms (plants, fungi, and many ciliated protists), there is a large vacuole but no obvious lysosomes. This is because vacuoles often perform the role of the lysosome in the cells that have them. There is quite a lot of variation in how this is arranged in organisms from different kingdoms, so we won’t say much about it, but it’s still important for you to know the relationship between the vacuole and the lysosome.
Lysosomes in TEM
Figure 04-30 shows an electron micrograph of three lysosomes in the cytosol of a ciliated unicellular protist. Lysosomes often contain what looks like strange bits and pieces. This material and the internal structure of the lysosome change over time because the contents are derived from the remains of cellular digestion. Sometimes the bits inside are very dark, and sometimes they’re not. This can make lysosomes a little bit difficult to identify in TEM, as their appearance changes depending on what they’re digesting.
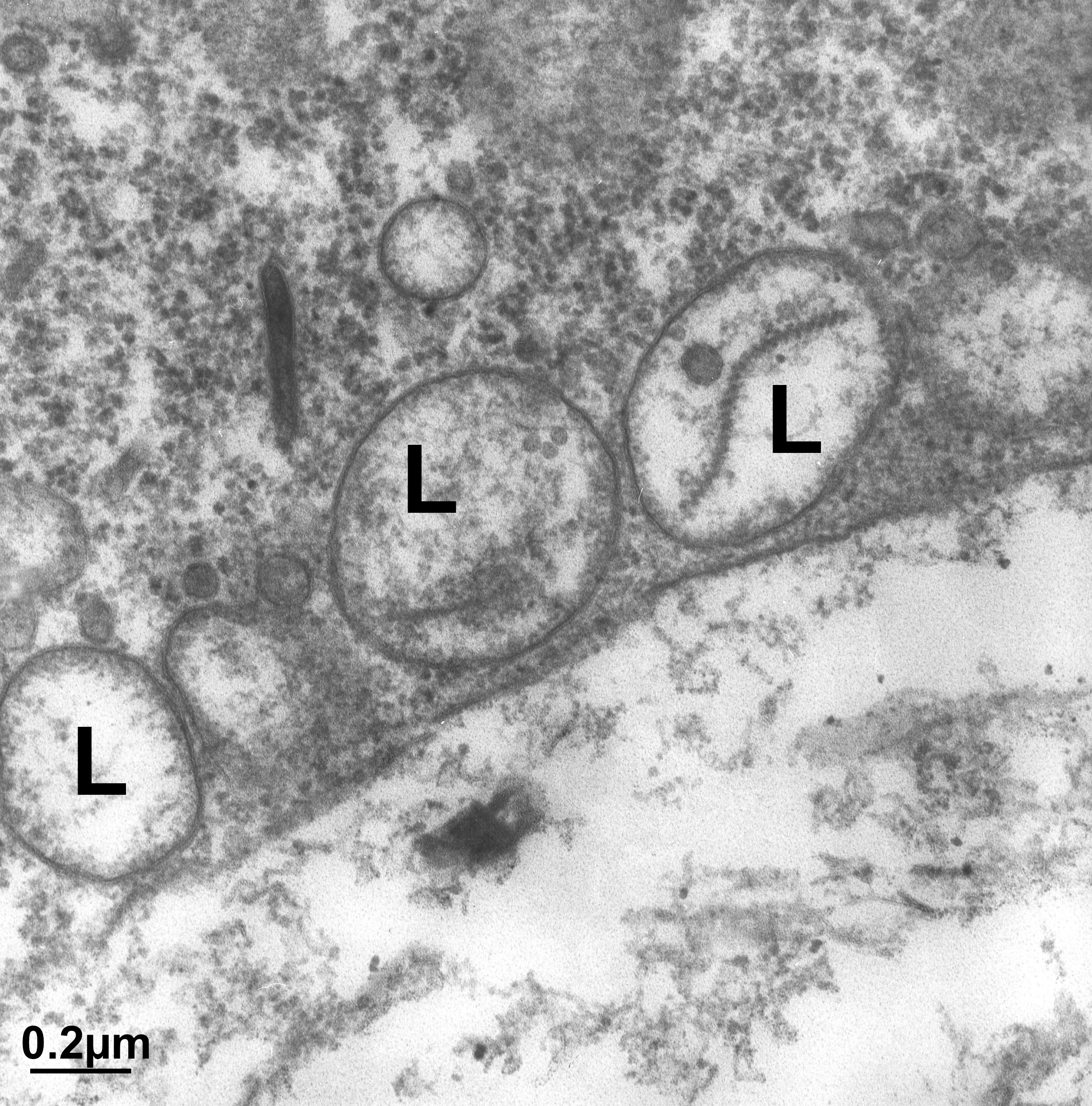
The Endocytic Pathway (a.k.a. Endocytosis)
The final pathway through the endomembrane system is the endocytic pathway, also known as endocytosis. This pathway starts at the plasma membrane instead of the ER. In this pathway, material is brought in from the exterior of the cell to the endosome. From the endosome, it can go several places:
- Proteins and other structures that are destined for degradation will go to the lysosome to be digested.
- Some cell-surface receptors are recycled back out to the plasma membrane after releasing their cargo in the endosome.
- There is some evidence, especially in plants, that some endocytic cargo gets sent to the TGN, where it may be sorted and redirected to another site afterward.
You can trace all these pathways in Figure 04-15, found in Topic 4.2.
Types of Endocytosis
When we discuss the endocytic pathway, we are discussing three distinct forms of internalization (Figure 04-31):
- Phagocytosis: the ingestion of large particles by cells. The particles are taken into phagosomes or food vacuoles. In multicellular organisms, this occurs primarily in specialized cells, like macrophages. However, aquatic single-celled protists, such as amoeba or paramecium, are also capable of this.
- Pinocytosis: the ingestion of fluid, ions, and other small particulates via small vesicles <150 nm in diameter. This is carried out by all cells and is believed to be nonspecific.
- Receptor-mediated endocytosis: the ingestion of specific molecules (e.g., cholesterol or iron) that are captured by cell-surface receptors to be packaged into vesicles.
We will look at each of these in turn.
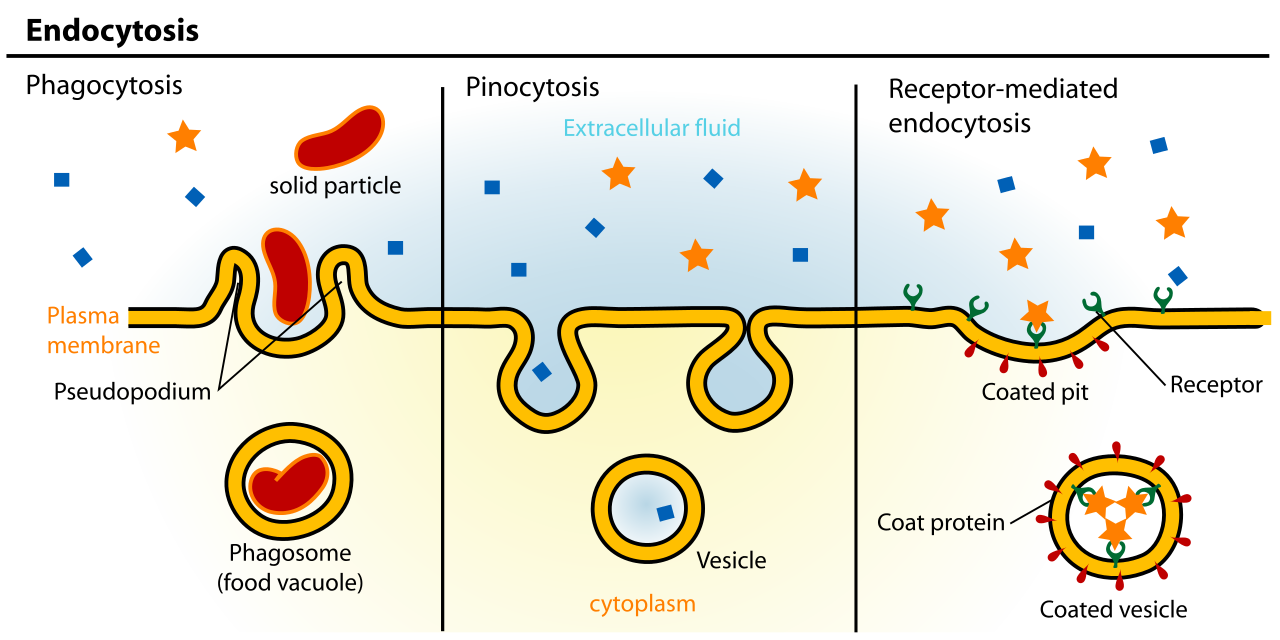
Phagocytosis
In phagocytosis, bacteria or other large particles are brought into the cell using large membrane-bound compartments we call phagosomes. Phagocytosis generally occurs as a response to the activation of cell-surface receptors, which results in the opening of calcium channels. The increased internal calcium concentration then signals the actin cytoskeleton to rearrange. The phagosome is then formed. Phagocytic cells play an important role in the removal of foreign organisms (like bacteria) and in the scavenging of dead or damaged cells in the immune system of many animals. Many of our white blood cells are capable of phagocytosis. In single-celled protists, this is also a mechanism to ingest food.
Phagosomes are complicated organelles to produce, as they require a great deal of membrane, which needs to be pulled up and around the structure that is being engulfed. Actin is thought to be heavily involved in this process, as it is a major mechanism used by the cell to push the membrane outward (more on this in Chapter 6), but the mechanism is still under investigation. Once the phagosome has formed, a lysosome will fuse with it to release digestive enzymes into the compartment, which can then be activated as the compartment acidifies. Interestingly, the acidic phagosome is also a site of active immune warfare in our bodies. Some bacteria are known to use the phagosome to invade their host. They are able to block lysosomal digestion and remain functional inside the phagosome. The bacteria that cause tuberculosis, Legionnaire’s disease, and listeria all manipulate the phagosome to protect themselves from the immune system in humans and other mammals.
Pinocytosis
This type of endocytosis can also be thought of as constitutive endocytosis, because it is thought to occur all of the time. Pinocytosis is relatively nonselective in what it brings into the cell (water, small ions, and other nutrients). It is also one of the ways that the cell manages to maintain a constant size despite the continuous flow of vesicles to the plasma membrane via secretion. For example, macrophages ingest about 25% of their own volume in fluid each hour and an area of membrane equal to its entire surface every 30 minutes. This is offset by the outward flow of vesicles (including membrane, fluid, and cargo) through exocytosis.
Pinocytosis may or may not use clathrin-coated vesicles for transport. More commonly, a special structure called a caveola is used. Caveolae (plural) are formed using a different protein complex that includes a protein named caveolin. They’re not as well understood as clathrin or the COP coats, so we won’t discuss them further.
Receptor-Mediated Endocytosis
Receptor-mediated endocytosis is a much more selective process than pinocytosis. It’s triggered by the binding of macromolecules to receptor proteins at the cell surface. The increased concentration of bound molecules triggers the formation of a vesicle. Endocytic cargo may be concentrated up to a thousandfold by binding and accumulating on the cell surface before incorporation into vesicles. Cargo molecules bind to receptor molecules that then bind to the adaptor protein (adaptin) and coat protein (clathrin in this case) that result in their concentration in the area of the forming membrane bud that will give rise to the endocytic vesicle.
An excellent example of this is the internalization of cholesterol. As you know from its chemical structure, cholesterol is not very soluble in aqueous environments, such as our blood, and this makes it difficult to transport from our digestive tract (where it’s absorbed from our food) to the rest of the cells in our body. As such, the cell packages cholesterol into a structure known as a low-density lipoprotein (LDL). The LDL can travel through the bloodstream and get endocytosed by the cells that require it (Figure 04-32).
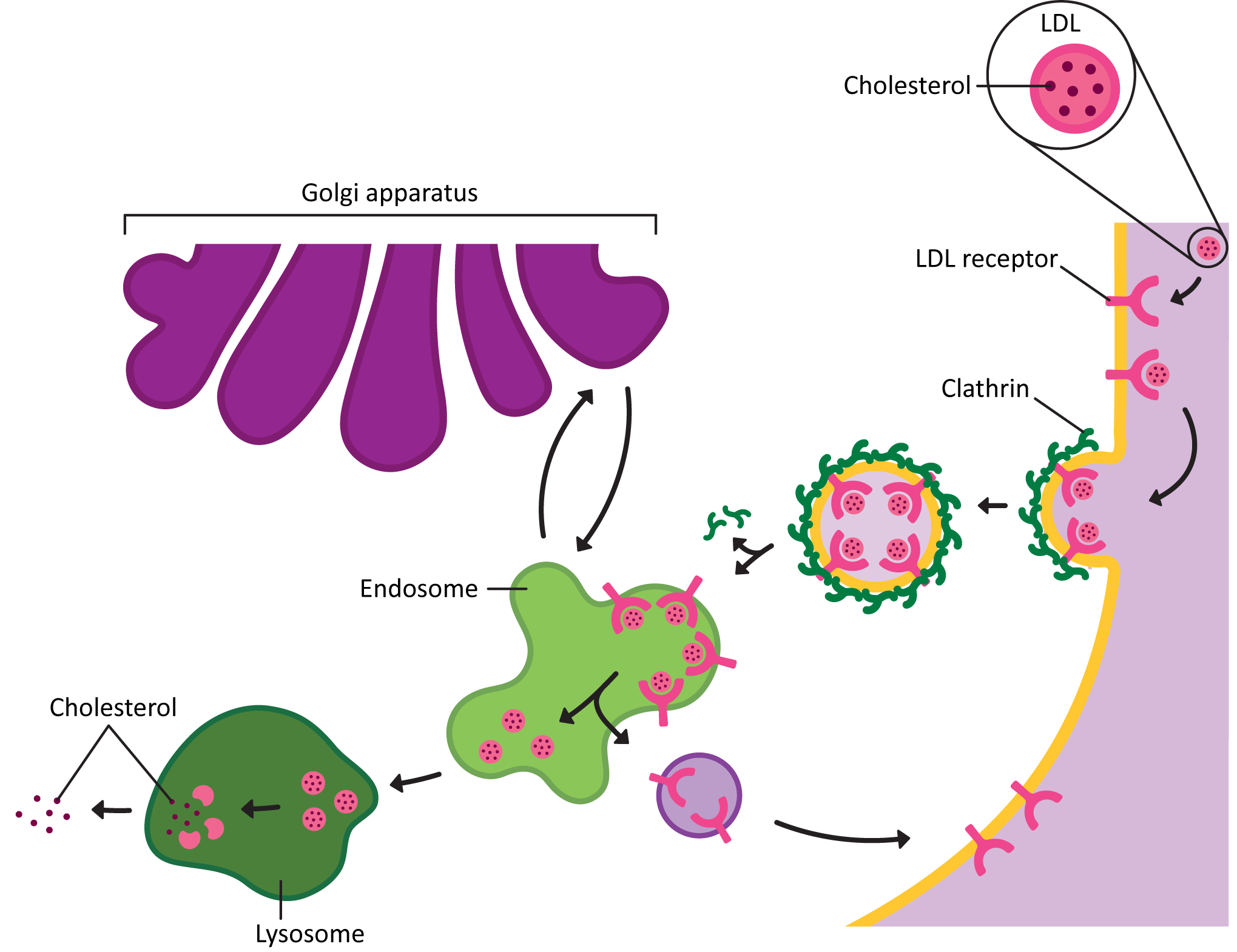
The following are the steps of receptor-mediated endocytosis of the LDL:
- The LDL complex binds to receptors on the cell surface, which triggers the formation of clathrin-coated vesicles.
- The vesicles then lose their coats and travel to and fuse with the endosome.
- In the acidic environment of the endosome, the LDL separates from its receptor.
- The LDL receptors are returned to the plasma membrane via transport vesicles. Cholesterol is released from LDL in the lysosome and available to use in membranes throughout the cell.
The Endosome Is Also a Sorting Center
The endosome is the second major sorting center in the endomembrane system (the first one being the TGN of the Golgi). It is the junction point between the lysosomal pathway and the endocytic pathway (Figure 04-32). This means that it must be able to differentiate between cargo that is headed to the lysosome and receptors that must be returned to either the TGN or the plasma membrane.
As if that were not complicated enough, endosomes receive receptors and cargo from the plasma membrane that may be headed to several different destinations:
- Many receptors are returned to the same plasma membrane domain from which they came, while their cargo is passed onto the lysosome. The LDL receptor is an example of this (shown in Figure 04-32).
- Some receptors are not recycled but instead sent to the lysosome to be degraded. This can be done with or without their cargo. Many growth hormones and receptors for irreversible developmental cues will follow this route.
- Finally, some receptors, along with their cargo, are transferred to another plasma membrane domain. This allows specific cargo to be passed from one side of the cell to another, in a process known as transcytosis. This is the process by which antibodies get passed from parent to child through the breast milk–producing cells when a child nurses.
Whether the cargo has the same fate as the receptor it rode in with or not is, in part, dependent on whether the acidic environment of the endosome causes cargo release. If the cargo is one that gets released by the acidic endosomal environment, then the cargo alone will end up in the lysosome, and the receptor is free to be recycled. Receptors that do not respond to low pH by releasing their cargo are more likely to travel with the cargo to the lysosome and get degraded.
Experimental Methods for Studying the Endomembrane System
As we have said many times in this chapter, the endomembrane system of a cell is essential, which means that the cell cannot survive without it. Essential cellular processes such as this create an interesting conundrum to researchers who wish to study them. One of the most common methods for studying a biological system is to disrupt it and study the effects. But when a system is essential, the cell dies if you disrupt it. Unsurprisingly, it is extremely difficult to learn much from a dead cell. Thus, scientists have had to get creative to find ways to study the secretory system in cells. Their methods have been very innovative and have been vital to our understanding of cell biology.
Visual Techniques—Microscopy
One of the most commonly used techniques these days to study how cargo moves through the endomembrane system is live-cell imaging of fluorescently labeled proteins. (Note: We have already spoken about visual techniques at length back in Chapter 1, so if you need a refresher, you may want to refer to the material found there.) We can combine live-cell imaging with other techniques described below to track how the system changes when mutations are made or blockages are temporarily induced.
As an example of the power of fluorescent microscopy to teach us about the endomembrane system, Video 04-08 shows the ER (green network) and the Golgi (purple blobs) in a plant leaf cell. In TEM we would see more detail of the structure, but since the samples must be dead and fixed, we cannot learn about the dynamics of these structures. In this live-cell video, we see the Golgi and ER both moving and changing shape dramatically throughout.
Biochemical Techniques
When researchers were first trying to figure out the endomembrane system, a common approach was literally to take it apart to investigate the job of each of these compartments in vitro (i.e., in a test tube). The biochemical techniques used were quite involved and complex. One common way to isolate different parts of the endomembrane system was to use a process called cell fractionation. In this process, the cells were literally put in a blender to break them up and then centrifuged at varying speeds, under different conditions, to separate rough ER from smooth ER from Golgi and so on. Researchers could then work with isolated membranes to see what each one was capable of on its own. In biology, isolating your “thing” of interest is a very common way of trying to understand function. Figure 04-33 shows an example of reconstructing an ER from isolated membranes in a test tube for an experiment.
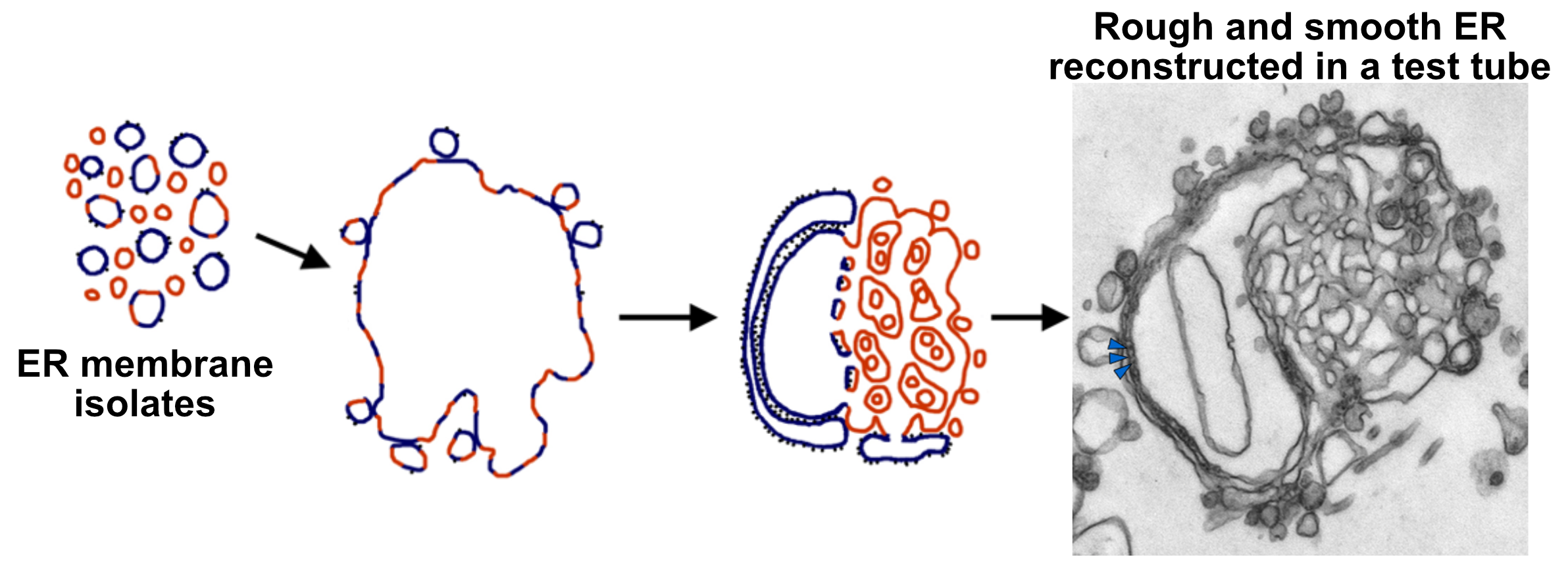
The Development of Temperature-Sensitive Secretion (sec) Mutants in Yeast and the Rise of Mutational Analysis as the Primary Tool for Studying the Endomembrane System
One of the most common ways for cell biologists to study cellular function is to disrupt a protein’s ability to function (through mutation of its gene) and then see what processes are disrupted when that protein is absent. When studying essential processes like secretion and the endomembrane system, this can pose a challenge. Many of the proteins involved are so important that the organism cannot survive at all without it. As you can imagine, this creates difficulties for scientists who want to better understand how the proteins of secretion function. The discovery of temperature-sensitive mutations was a turning point in our understanding of many essential cellular processes—so much so that the combination of temperature-sensitive mutations and live-cell imaging techniques is a very large part of how endomembrane research is done today.
A temperature-sensitive mutation is one in which the protein amino acid sequence has changed due to the mutation, but it’s not so severe a change that the protein is nonfunctional. The protein is still able to fold more or less correctly. At certain temperatures (known as the permissive temperature, usually a few degrees lower), these proteins work just fine. They are stable and are capable of doing their job. However, if the temperature is changed even a few degrees (to what is known as the restrictive temperature), the protein becomes unstable, denatures, and is no longer able to maintain its function. As long as the cell is kept at the restrictive temperature, the protein will not regain its function. If left for long enough, the cell will die. If the cell is moved back to the permissive temperature, in time, the protein will renature, regain function, and thus the regular cellular processes will resume.
An excellent example of a temperature-sensitive mutation is that of the coloring of a Siamese cat (Figure 04-34). Siamese cats very famously have lighter beige hair on their bodies and darker brown hair on their extremities (paws, face, tail). The particular coat coloring of a Siamese cat is the result of a temperature-sensitive mutation in the tyrosinase enzyme, which is one of the many enzymes involved in producing the brown color pigment, melanin. The cat’s extremities are slightly cooler, so in these areas, the tyrosinase enzyme is functional, and the dark color is produced. Near the animal’s core, the temperature is a little bit higher, so the enzyme does not function, melanin is not produced, and the animal has albino fur.

One of the advantages of this system, from a cell biology perspective, is that a temperature-sensitive mutation is heritable, because a cell at a permissive temperature can survive long enough to divide and/or reproduce. This is important in experiments, as it is very challenging to generate these cell lines with mutations in specific genes. Even if one does regenerate the same mutated gene, there is the possibility that the same gene with a different mutation will not show the same effects, so being able to maintain a cell line with a specific mutation in an essential gene like the ones of the endomembrane system allows us to spend more time studying each mutation and thus learn more about cell biology as a whole.
For the endomembrane system, a great deal of work has been done on the budding yeast, Saccharomyces cerevisiae. There are multiple reasons why they were an ideal system in which to study secretion:
- They are single-celled Eukaryotes with a very short life cycle (~1–2 hours) that tolerate a wide range of temperatures. This makes them ideal for creating temperature-sensitive mutants and studying the effects.
- They have a small genome with very few genes compared to us (~12 million base pairs and ~6,200 genes). A simpler system is easier to study.
As such, a great deal of work has been done in budding yeast in order to study the endomembrane system and secretion. A great many temperature-sensitive mutants exist that have shed light on many different parts of the protein machinery that drives function. They are called the secretion, or Sec, mutants. One such example is Figure 04-35, where we see the difference in one of the mutants, called n-Sec1, as restrictive and permissive temperature.
Chapter Summary
The endomembrane system is a dynamic system involving many organelles, each with a unique job to do. Fittingly, the regulation of this system is equally dynamic and complex. In this chapter, we have discussed the roles of the ER, Golgi, endosomes, and lysosome and how resident and transient proteins are sent to or pass through these different compartments.
The ER is the entry point into the endomembrane, and proteins targeted anywhere in the endomembrane system contain an ER insertion sequence that is part of their primary sequence. Depending on the type and location of these ER targeting sequences, the protein can be either soluble or membrane bound. This starts their journey in the endomembrane system.
Proteins move to different organelles within the endomembrane system via a small membrane-bound vesicle. These vesicles provide specific transport based on the proteins contained in each vesicle and the targeting machinery on the surface.
The Golgi is the protein processing and sorting center. All proteins that enter into the endomembrane system pass through this hub. It is the primary site of glycosylation, and it is the primary site of production of cell walls in plants, fungi, and algae. Proteins can reach the cell exterior, be sent back to the ER, or be targeted to the endosome/lysosome from the Golgi.
Finally, a secondary sorting hub, known as the endosome, receives cargo from the cell surface and redirects it. Proteins and other molecules can be sorted and returned to the Golgi / cell surface or sent to the lysosome to be degraded.
This is a crucial system that is also quite dynamic, which makes it difficult to study. However, some pivotal experimentation as well as advances in technology have allowed scientists to gain significant insights into this highly intricate system. This chapter simply scratches the surface of the scientific knowledge on the endomembrane system and we hope that you find it as interesting as we do. If not, we hope that you have at least a deeper appreciation for the importance and complexity.
Review Questions
Note on usage of these questions: Some of these questions are designed to help you tease out important information within the text. Others are there to help you go beyond the text and begin to practice important skills that are required to be a successful cell biologist. We recommend using them as part of your study routine. We have found them to be especially useful as talking points to work through in group study sessions.
Topic 4.1: The Endoplasmic Reticulum
- Define the following terms and determine which ones might be synonyms of each other:
- targeting sequence
- N-terminal START sequence
- signal recognition particle
- signal recognition particle receptor
- signal peptidase
- START transfer sequence
- STOP transfer sequence
- Describe the differences between smooth, rough, and transitional ER.
- How come disulfide bridges are more common in secreted proteins than in cytosolic proteins?
- Define the role of chaperones in protein processing. What determines whether a given protein will need chaperones for proper folding?
- How many ways can a given protein fold? What determines the folding pathway that a given protein takes?
- What happens to proteins that fail to fold properly in the ER?
- Explain the idea of a cellular compartment, the ideas of specificity of content and function, and the “sidedness” of membranes. How does the cell maintain the proper “side” of the membrane exposed to the cytosol in all organelles (including the plasma membrane)?
- Outline the mechanisms by which proteins are transported to different membrane-bound compartments in the cell.
- What are the common properties of “START transfer” and “STOP transfer” sequences?
- What determines whether the N-terminal end of a protein is in the cytosol or inside the ER cisterna?
- Describe the general steps to protein insertion into the ER.
- Many membrane proteins contain a number of membrane-crossing domains. How do these proteins get inserted into the membrane?
- Name a few examples of proteins that might be ER resident proteins and how they are able to stay in the ER.
- Compare and contrast the mechanism of protein insertion in the ER with nuclear import. Think about the ideas of necessary and sufficient and how they apply to the endomembrane protein targeting system.
Topic 4.2: Vesicle Transport
- Make a table of the different coat proteins, and include all of the protein components for each one (coat, adaptor, GTPase, and whether it needs a separate scission protein, location in the cell, and/or trafficking pathways)
- What is the role of clathrin or other coat proteins in the selection of cargo and the formation of membrane vesicles?
- How do coat proteins facilitate the process of vesicle formation?
- What sort of forces need to be overcome to form a vesicle?
- How do Rabs and tethers facilitate vesicle docking?
- What are SNAREs, and how do they regulate membrane fusion?
- Sometimes the enzymes of the ER are accidentally transported to the Golgi. How are these proteins recovered and returned to the ER?
Topic 4.3: The Golgi Apparatus
- Define the following terms and determine which ones might be synonyms of each other:
- cis cisternae
- medial cisternae
- trans cisternae
- trans Golgi network
- lumen
- glycoprotein
- proteoglycan
- KDEL receptor
- What portion of a membrane glycoprotein is glycosylated (cytosolic, membrane-crossing domain, or the portion facing the lumen of the ER)?
- What is the role of glycolipids in the glycosylation of proteins?
- How do cells “know” where to add oligosaccharides to prospective glycoproteins during the glycosylation process?
- The lumen of the ER contains a number of enzymes that are involved in protein processing. These proteins are retained in the ER lumen, even though most of the material in the lumen is transferred to the Golgi through vesicle transport. How does this happen?
- Compare and contrast the structure and function of plant and animal Golgi.
- Explain the central role of the Golgi in the endomembrane system with respect to protein synthesis, import of material from the cell exterior, vesicle traffic, and cellular digestion.
Topic 4.4: Pathways Through the Endomembrane System
- Describe the structure and function of the lysosome. How does it fit into the rest of the endomembrane system?
- Compare and contrast the regulated and constitutive secretory pathways with regard to the types of material carried and the regulation of secretion.
- Trace a molecule of a lysosomal protein from its point of synthesis on a ribosome in the cytosol to its final destination in a lysosome. Include everything that must happen to it for successful targeting to the lysosome and where each of those things happen in the cell. Be as specific as you can.
- Explain how the mannose-6-phosphate receptor is recycled.
- What determines whether a protein is sorted into the regulated or the constitutive secretory pathway?
- What is the difference between early and late endosomes?
- Define receptor-mediated endocytosis and describe the steps. Be as specific as you can.
- In both endocytosis and secretion, proteins are sorted into vesicles destined for different target compartments. Where does sorting occur in each pathway? What are the similarities and the differences?
- How does an early endosome “become” a late endosome, and then a lysosome?
- Do all receptors that get endocytosed get recycled back to the plasma membrane? What happens to the ones that don’t?
- Describe the common experimental techniques to study the endomembrane system along with the pros and cons of each one.
References
Harrison, S., & Kirchhausen, T. (2010, July). Conservation in vesicle coats. Nature, 466, 1048–1049. https://doi.org/10.1038/4661048a
Kang, B. H., Nielsen, E., Preuss, M. L., Mastronarde, D., & Staehelin, L. A. (2011). Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic (Copenhagen, Denmark), 12(3), 313–329. https://doi.org/10.1111/j.1600-0854.2010.01146.x
Noble, A. J., & Stagg, S. M. (2015). COPI gets a fancy new coat: An interconnected scaffolding of proteins bends the membrane to form vesicles. Science, 349(6244), 142–143. http://www.jstor.org/stable/24748469
Novick, P., & Schekman, R. (1979). Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America, 76(4), 1858–1862. https://doi.org/10.1073/pnas.76.4.1858
Staehelin, L. A., & Kang, B. H. (2008). Nanoscale architecture of endoplasmic reticulum export sites and of Golgi membranes as determined by electron tomography. Plant Physiology, 147(4), 1454–1468, https://doi.org/10.1104/pp.108.120618
Voelz, V. A., Jäger, M., Yao, S., Chen. Y., Zhu, L., Waldauer, S. A., Bowman, G. R., Friedrichs, M., Bakajin, O., Lapidus, L. J., Weiss, S., & Pande, V. S. (2012). Slow unfolded-state structuring in Acyl-CoA binding protein folding revealed by simulation and experiment. Journal of the American Chemical Society, 134(30), 12565–12577. https://doi.org/10.1021/ja302528z
Weidman, P. J. (1995). Anterograde transport through the Golgi complex: Do Golgi tubules hold the key? Trends in Cell Biology, 5(8), 302–305. https://doi.org/10.1016/s0962-8924(00)89046-7
Yuen, C. Y. L., Wang, P., Kang, B. H., Matsumoto, K., & Christopher, D. A. (2017). A non-classical member of the protein disulfide isomerase family, PDI7 of Arabidopsis thaliana, localizes to the cis-Golgi and endoplasmic reticulum membranes. Plant and Cell Physiology, 58(6), 1103–1117, https://doi.org/10.1093/pcp/pcx057
“The transportation system of the eukaryotic cell… [that] has many other important functions such as protein folding. It is a type of organelle made up of two subunits—rough endoplasmic reticulum (RER), and smooth endoplasmic reticulum (SER). The endoplasmic reticulum is found in most eukaryotic cells and forms an interconnected network of flattened, membrane-enclosed sacs known as cisternae (in the RER), and tubular structures in the SER.” Source: Endoplasmic reticulum. (2023, June 12). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Endoplasmic_reticulum
A compartment at the heart of the endomembrane system responsible for sorting and processing proteins in that system.
A roughly spherical membrane-bound organelle that contains digestive enzymes. This compartment breaks down materials delivered from the endocytic pathway.
Intracellular sorting centers that are the first location molecules are delivered to once they are endocytosed. There are two classifications of endosomes, early and late, which are marked by different cellular markers. Some vesicles from the TGN can be routed to the endosome as well. Endosomes sort materials that need to go to the lysosome from those that need to go back to the Golgi or cell membrane.
A small membrane-bound structure containing cargo proteins destined for a particular cellular location in the endomembrane system.
Refers to proteins leaving the cell through the secretory pathway.
Refers to the protein trafficking pathway originating at the ER and ending at the plasma membrane, where soluble proteins are released into the extracellular space and integral membrane proteins are embedded in the plasma membrane. Proteins are first imported into the ER and moved to the Golgi for further processing before being sent to the plasma membrane / cell exterior. This is the default pathway for proteins targeted to the ER and does not require additional targeting sequences beyond ER insertion sequences.
Refers to the vesicle trafficking pathway originating at the TGN and ending at the lysosome for lysosomal resident proteins. Proteins are recognized by a receptor at the TGN and transported to the endosome where the protein is disassociated from the receptor. The protein then travels to the lysosome. The receptor is transported back to the TGN for further rounds of trafficking.
The pathway by which materials are brought into the cell. Once materials are in the cell, they transit to the endosome and then to the lysosome.
The process in which outside materials and cargo are brought into the cell after passing through the membrane and forming a vesicle once inside. Endocytosis includes pinocytosis, phagocytosis, and receptor-mediated endocytosis.
The process of modifying proteins after they are translated.
“The reaction in which a carbohydrate (or ‘glycan’), i.e., a glycosyl donor, is attached to a hydroxyl or other functional group of another molecule.” Source: Glycosylation. (2023, June 21). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Glycosylation
“Proteins that assist the conformational folding or unfolding of large proteins or macromolecular protein complexes.” Source: Chaperone (protein). (2023, April 2). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Chaperone_(protein)
“A protein coplex that is responsible for degrading unneeded or damaged proteins through proteolytic chemical reactions that break peptide bonds.” Source: Proteasome. (2023, January 29). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Proteasome
“A small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms.” Source: Ubiquitin. (2023, July 18). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Ubiquitin
“The breakdown of proteins into smaller polypeptides or amino acids.” Source: Proteolysis. (2023, May 10). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Proteolysis
The covalent linkage of thiol groups between two thiol residues. Commonly, these are found on the R groups of cysteine amino acids.
The interior fluid of the endoplasmic reticulum (ER).
“A flattened membrane vesicle found in the endoplasmic reticulum and Golgi apparatus. Cisternae are an integral part of the packaging and modification processes of proteins occurring in the Golgi.” Source: Cisterna. (2023, January 3). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Cisterna
A section of the ER where the membrane is studded with ribosomes and is responsible for the process of protein production and protein folding.
A region of the ER that does not contain any ribosomes. It is involved in many metabolic processes, including phospholipid synthesis, needed for membrane formation.
A region of ER where the rER and sER meet. Often it is the exit site for vesicles leaving the ER.
The site on the endoplasmic reticulum (ER) where vesicles accumulate and bud off bound for the Golgi.
A cellular response to unfolded proteins whereby they are targeted for destruction.
A protein that is not embedded in a membrane and can diffuse freely in an aqueous environment.
A type of protein that is embedded into biological membranes. Transmembrane proteins are an example of integral membrane proteins that span the entire biological membrane and are used to transport material from one side to the other.
A sequence of hydrophobic amino acids (approx. 8–10) that serve as a binding site for SRP. This triggers its cotranslational insertion into the ER. There are several variations of this sequence depending on the location in the protein’s sequence, including N-terminal START sequence, internal START sequence, and internal STOP sequence.
A sequence of hydrophobic amino acids located at the N-terminus of a protein. This sequence will be cleaved after the protein has been imported into the ER.
A start signal sequence that is somewhere internal on the protein as opposed to at the N-terminus of the protein. The START sequence is always either the first ER insertion sequence or one that directly follows a START (if there are multiple ER insertion sequences). Unlike the N-terminal START sequence, this is not cleaved off after the transfer of the protein has been completed.
An ER insertion sequence that is located after a START sequence. It signifies to the translocation channel to stop threading the translated protein into the ER lumen. This sequence eventually is released into the membrane and exists as a transmembrane domain for the protein.
“An abundant, cytosolic, universally conserved ribonucleoprotein (protein-RNA complex) that recognizes and targets specific proteins to the endoplasmic reticulum in eukaryotes.” Source: Signal recognition particle. (2022, October 18). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Signal_recognition_particle
Channel necessary for inserted proteins into the ER. When an ER insertion sequence emerges from a protein, SRP binds and brings it to the SRP receptor, which interacts with the translocation channel. Anything after this sequence is threaded through the channel in the ER lumen unless it runs into a STOP translocation sequence.
An enzyme that is responsible for cleaving the N-terminal start sequences from the rest of the protein once trafficked and inserted into the ER.
“Refers to proteins that remain in the endoplasmic reticulum, or ER, after folding has finished and are not exported to other organelles.” Source: ER resident proteins. (n.d.). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Endoplasmic_reticulum_resident_protein
A targeting sequence defined by the amino acids KDEL (lysine, aspartate, glutamate, and leucine). If these proteins travel to the Golgi, this sequence helps them to be recognized by a receptor to be returned to the ER.
A targeting sequence defined by the amino acids KDEL (lysine, aspartate, glutamate, and leucine). If these proteins travel to the Golgi, this sequence helps them to be recognized by a receptor to be returned to the ER.
“A region of a protein’s polypeptide chain that is self-stabilizing and that folds independently from the rest.” Source: Protein domain. (2023, June 4). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Protein_domain
A schematic representation of the protein sequence with highlighted areas indicating protein domains (regions of the protein with a particular function).
The process of a vesicle forming from the donor membrane in a cell or organelle.
The movement of proteins and other biological molecules through the cells in membrane compartments called vesicles.
Involves the use of tether proteins to get closer to the target compartment prior to fusion.
The process of merging a vesicle membrane with the membrane of the target compartment, thereby releasing the contents into the lumen of the target (or the extracellular space in the case of the cell membrane). The process is mediated by SNARE proteins.
Responsible for binding to specific forms of cargo like proteins. Cargo receptors help the cargo get loaded into the correct vesicle for transport.
Proteins that play an important role in vesicle formation, where they help to gather scaffold proteins that are needed to form the clathrin coat of the vesicle.
Help support the formation of a budding transport vesicle. Coat proteins bind to adaptor proteins and cargo receptors to ensure that the proper cargo is recruited into these structures.
Enzyme that functions as a molecular switch. GTPases bind to GTP and GDP. Depending on their bound state, they will have different functions. This is a large class of proteins with a wide variety of functions.
Proteins that are carried within vesicles to a particular location. These can be proteins that are secreted but also other resident proteins that need to travel to a particular destination in the endomembrane system.
A term used to describe the process of vesicles detaching from the donor compartment.
Transport vesicles that contain a coat with clathrin triskelions. These coated vesicles participate in a variety of pathways, notably endocytosis and the lysosomal targeting pathway.
The end location of the Golgi where proteins are sorted into vesicles bound for their next destination. Primary post-Golgi locations include the plasma membrane and the endosome.
Transport vesicles that contain a coat with COPI units. These coated vesicles participate in a variety of pathways, notably Golgi to ER (retrograde pathway).
Vesicle tracking that moves opposite of the default movement of traffic. In the secretory pathway, an example would be going from the Golgi back to the ER.
Transport vesicles that contain a coat with COPII units. These coated vesicles participate in a variety of pathways, notably ER to Golgi (anterograde pathway).
Involves movement of coated vesicles in the “forward” direction. Depending on the trafficking pathway, this can be different destinations.
The name for clathrin monomers that form a cage-like structure around a forming transport vesicle.
Part of a large family of enzymes known as GTPases that play a crucial role in endocytosis, cytokinesis, scission of newly formed vesicles, and division of organelles.
A protein that binds to the KDEL amino acid sequences in the Golgi to help load them into vesicles back to the ER.
A process in which cargo from outside the cell enters, creating inward budding vesicles from the plasma membrane. This process is controlled by receptors located on the cell surface, and cargo is only allowed to enter after a ligand has bonded to a corresponding receptor.
An endocytic process that occurs continually and nonselectively. The membrane invaginates, bringing in small molecules from the external environment.
A small protein that is a member of the GTPase family and is important in the processes of vesicle trafficking.
Tether proteins in the target membrane help with vesicle docking. Tether proteins bind to the Rab proteins to bring the vesicle closer to the target membrane such that SNARE proteins are then able to bind and promote vesicle fusion.
Part of a class of proteins needed for vesicle fusion. When a vesicle nears a target membrane, if there are appropriately matched SNARE proteins, they will bind. This binding causes the proteins to wind up, physically bringing the membranes in close proximity such that the lipid bilayers can fuse.
A type of SNARE located in the membrane of transport vesicles.
A type of SNARE that is located in the membranes of target membranes and will pair with specific v-SNAREs found on vesicle membranes.
The area inside a membrane-bound organelle. For instance, the ER lumen, Golgi lumen, and lysosomal lumen.
The region between the two membranes of the nuclear envelope.
A model to describe the movement of proteins through the Golgi apparatus. In this model, each cisterna matures into the next. This means that the cis cisternae become the medial, then the trans cisternae, which eventually turn into vesicles in the trans Golgi network (TGN) for further trafficking.
A model describing the function of the Golgi. In this model, the cisternae stay stable in a Golgi. Proteins traveling through this system will bud from each cisterna and fuse with the next subcompartment using vesicles.
“The attachment of an oligosaccharide, a carbohydrate consisting of several sugar molecules, sometimes also referred to as glycan, to a nitrogen atom (the amide nitrogen of an asparagine [Asn] residue of a protein), in a process called N-glycosylation.” Source: N-linked glycosylation. (2023, July 9). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/N-linked_glycosylation
“The attachment of a sugar molecule to the oxygen atom of serine (Ser) or threonine (Thr) residues in a protein.” Source: O-linked glycosylation. (2021, October 19). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/O-linked_glycosylation
“Transmembrane lipid transporter proteins located in the membrane that belong to ABC transporter or P4-type ATPase families. They are responsible for aiding the movement of phospholipid molecules between the two leaflets that compose a cell’s membrane (transverse diffusion, also known as a ‘flip-flop’ transition).” Source: Flippase. (n.d.). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Flippase
An enzyme that adds a carbohydrate group (‘glycan’) to a biological molecule.
A special protein structure where cellulose is made in plant cells destined for use in the cell wall.
A form of secretion that is regulated by external factors like signals that allow certain secretory vesicles to fuse with the plasma membrane.
The process of continuous secretion of molecules from the cell or organelle that is not dependent on external factors. This is also considered to be the default secretory pathway.
When referring to proteins, it means that they cluster together into clumps. Oftentimes these clumps are no longer functional.
Proteins that form clusters with other proteins. The aggregation tends to be composed of misfolded proteins that join to other misfolded proteins.
An organelle present in plants and fungi that is responsible for the degradation of materials originating in the endocytic pathway. Vacuoles also play a storage role that is not present in the animal lysosome, which is the functional equivalent in animal cells.
“A vesicle formed around a particle engulfed by a phagocyte via phagocytosis.” Source: Phagosome. (2023, February 11). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Phagosome
A mannose sugar phosphorylated on the sixth carbon with a phosphate.
The protein receptor that recognizes M6P on proteins.
The process of bringing in a large particle (like a bacterium or a cancerous cell) for digestion.
“A type of transcellular transport in which various macromolecules are transported across the interior of a cell. Macromolecules are captured in vesicles on one side of the cell, drawn across the cell, and ejected on the other side.” Source: Transcytosis. (2022, September 25). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Transcytosis
“The process used to separate cellular components while preserving individual functions of each component.” Source: Cell fractionation. (2023, February 22). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Cell_fractionation
Mutants of a gene where the result is a lack of activity when exposed to its restrictive temperatures. Often proper function is restored when brought back to the optimal (permissive) temperature.

