7 Cell Signaling
Introduction
In order to survive, a cell must be able to understand its environment. This is true whether the cell is a single-celled organism or part of a larger, more complex multicellular organism. Cells communicate with their environment through a process called signaling. Cell signaling is how the cell collects information and then responds with an action at the correct time. Signaling is the initial event associated with many key cellular functions, from the correct timing of cell division, to the decision to migrate in a particular direction, and even to whether a cell needs to go through programmed cell death. So many of the cellular events we explore in biology are dependent on signaling to happen correctly. Not only that, but many of the concepts covered in upper-level biology courses are, at their hearts, studies of how the cell receives information and responds to it. For example, developmental biology, sensory perception, endocrinology, and even physiology will make much more sense if you have a foundational understanding of how signaling works.
Thus, our focus in this chapter is to dissect the process of signaling and take a look at the parts of a signaling cascade and some of the commonly recurring themes and patterns. That way, when you encounter any pathway in any context, you will have the ability to “read it” through and identify the patterns within it to correctly interpret the outcomes of that pathway.
Topic 7.1: General Principles of Signaling
Learning Goals
- Explain the three stages of a general signaling cascade.
- Provide examples of common protein families involved in each stage.
- List and explain the different types of intracellular responses possible for a given signal.
- Distinguish among relay, amplification, integration, and distribution steps in a signaling cascade.
- Correctly interpret schematic representations of signaling cascades.
While signaling is a beautiful coordination of cellular events, it also is a very complex process. It can be very confusing to keep track of what the cell is doing at any given time due to the sheer number of signals, the overlap in signaling pathways, and sometimes even the competing signaling events occurring simultaneously. Additionally, a wide range of cellular “behaviors” are mediated through a small set of extracellular signals. Thus, the way that a cell responds to a specific extracellular signal will depend on what genes are being expressed in the cell at that specific moment in time. For example, acetylcholine is an extracellular signal that has different effects in different cell types. It is released at a neuromuscular junction by neurons to promote muscle contraction. In cardiac pacemaker cells, it signals that the heart rate should lower, so the pacemaker cells fire at a decreased rate. Finally, salivary glands also respond to acetylcholine by increasing the synthesis and secretion of saliva by the endomembrane system. Each of these scenarios is the result of different modes of release of the acetylcholine, different receptors receiving it on the cell surface, and also differential expression of the internal components of the acetylcholine pathway.
Our focus for this topic is to lay some groundwork for understanding signaling by looking at the general principles that underlie cell signaling. Then in the next topic, we’ll look at some examples of actual mechanisms that are commonly used in cell signaling. These tools should allow you to interpret any signaling pathway in the future and begin to understand how they mediate a response.
The video below (Video 07-01) gives a quick overview of the mechanism of cell signaling and the terminology you will need to understand this process.
Types of Cell Signaling
Before we start dissecting the various components of a signaling cascade, we must first understand the different forms of signaling that can occur. There are five different types of signaling that are common in cells: endocrine, neuronal, paracrine, autocrine, and juxtacrine (Figures 07-01 and 07-02, as well as Video 07-01). A multicellular organism (such as ourselves) is likely to use most or all of these different forms in different tissues/scenarios. Comparatively, single-celled organisms are more limited in their options, since they do not specialize, nor do they require communication as frequently between the different cells.
The different types of signaling modes differ in both the distance the signal travels and the speed with which it travels from the site of signal release to the site of receptor binding. They will also differ in the affinity that the receptor will have for the signaling molecule (also known as a ligand). The affinity of a ligand for its receptor refers to how “well” the ligand binds to the receptor. A receptor that is regularly flooded with a high concentration of ligands will not need to be as sensitive as one that must find a single molecule in the vast sea of the extracellular environment. Receptors that are more sensitive are said to be high affinity compared to those that are less sensitive to the presence of their ligand. It is also true that high-affinity receptors bind more tightly to their ligands and are less likely to uncouple after binding than lower affinity receptors.
Long-Distance Signaling: Endocrine (Slow) and Neuronal (Fast) Signaling
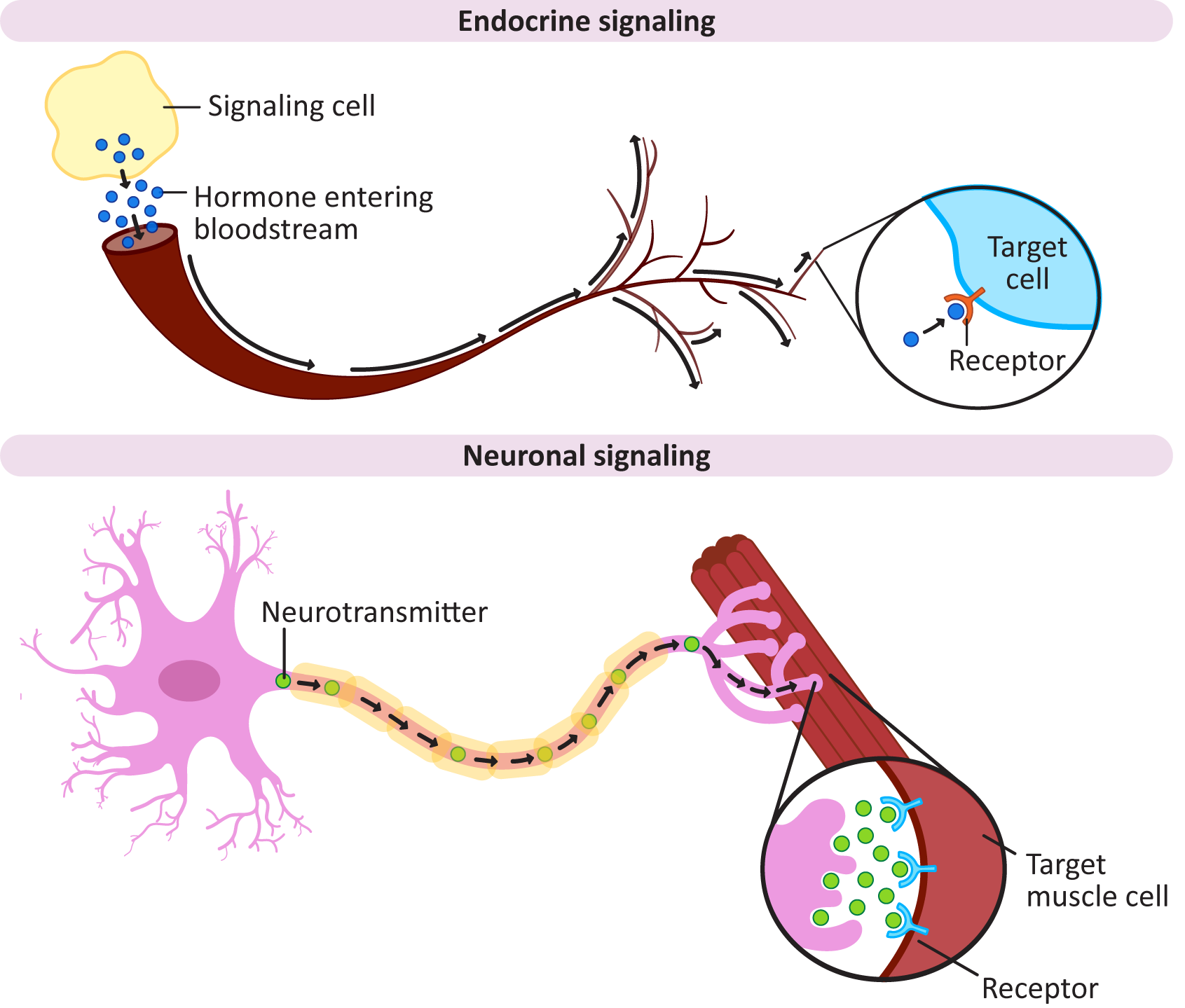
Signaling over long distances can be challenging, as there may be hundreds, if not thousands, of cells between the source of the signal and the cell that must receive it. Both plants and animals are able to transport signals over long distances; however, animals are the true experts at long-distance signaling, with both a fast and a slow mechanism to do so. Plants, on the other hand, mostly focus on short-range signaling. They have very few options for long-distance signaling, none of which are considered to be terribly fast. Endocrine and neuronal signaling are illustrated in Figure 07-01.
Endocrine signaling uses hormones, which are sent throughout the organism, as the initial signaling molecule. In animals, this is done via the bloodstream. Using the bloodstream for transport allows the signal to be produced at a single site in the organism (e.g., the adrenal glands), but it can have an effect in many places that are far away:
- Endocrine signaling is considered to be slooow, as the signal must be produced and secreted into the bloodstream, moved throughout the body, and then picked up by another cell that is likely very far from the site of ligand release. It can take minutes for the signal to be received, which is quite a long time in the world of cells and signaling.
- On the other hand, since the hormone will become very dilute as it moves through the body and the receptor must be able to find and bind the ligand even in these low-concentration conditions, the receptors for endocrine signals are quite sensitive. In some cases, a single molecule from the bloodstream can be detected. A receptor that can be activated by a small, dilute dose of signal is said to have high affinity for its ligand.
- Examples of endocrine signaling ligands include most hormones (i.e., insulin, adrenaline, estrogen, growth hormones, etc.).
Neuronal signaling is the type of signaling used by the nervous system. In a nutshell, an electrochemical signal is sent through our neurons, across large distances, to elicit a response:
- This kind of signaling is quite fast, which is good, considering that neuronal signaling is what makes you move your hand when you touch a hot surface by accident. The signal is passed along and received in a matter of milliseconds.
- Quite a bit of this process happens within the nerve cell. Nerves are very long cells—as an example, your sciatic nerve starts at the base of your spine and ends in your foot and is about 1 m long! Keeping the signal inside the cell for as long as possible makes it much easier for the signal to move quickly.
- At some point the signal will need to exit the neuron and be passed to the muscle or other tissue that is expected to respond. Multiple chemical signals are used to help with this part. Collectively, we call these signals neurotransmitters. You likely have heard of many of them: dopamine, epinephrine (also known as adrenaline), and acetylcholine, for example. Some recreational drugs act by affecting the ability of neurons to send and/or receive neurotransmitter signals.
- Neurotransmitters help the signal move from the neuron to the target cell (like a muscle cell). The gap between the nerve cell and the muscle cell is really small, so the signal doesn’t have to diffuse very far. To increase the chances that the signal will be received as quickly as possible, the nerve cell floods the entire area with the neurotransmitter. As such, these receptors are not as sensitive as those used for endocrine signaling…they don’t really have to be. Thus, we consider the receptors in the synapse to be low affinity.
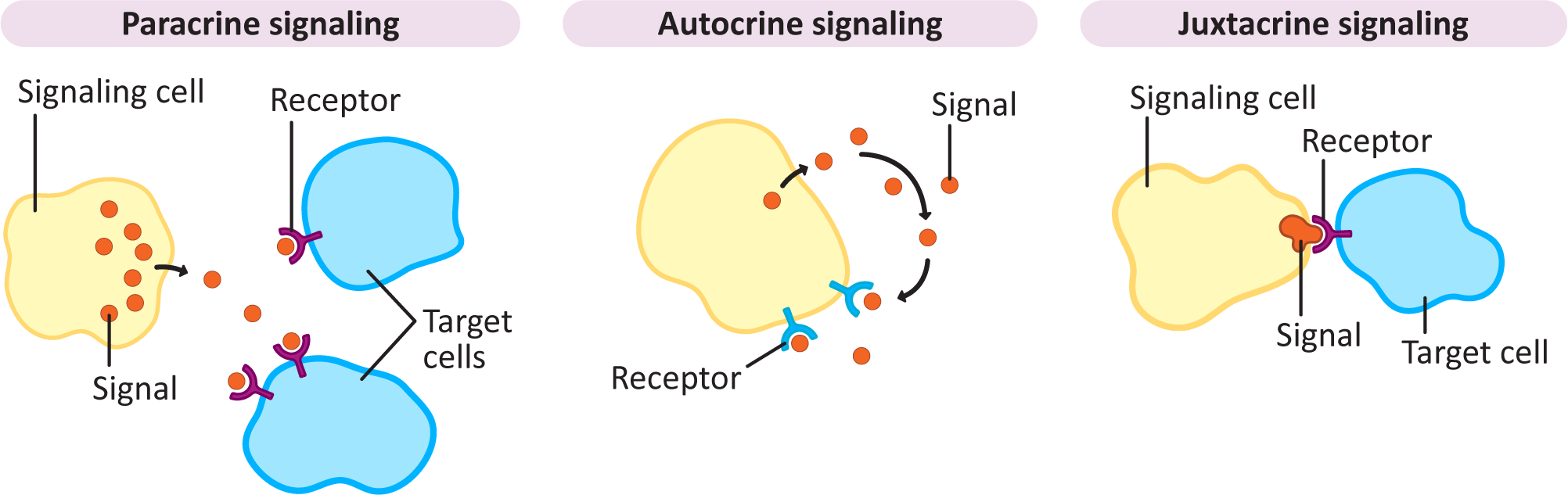
Medium- to Short-Distance Signaling: Paracrine (Diffusion Based), Juxtacrine (Contact Dependent), and Autocrine (Self-)Signaling
In many ways, signaling at closer range is much easier than long-distance signaling, as the signal can be released into the extracellular space and simply left to diffuse. This is one of the most common types of signaling in development. Plants, algae, and fungi don’t really have any long-distance options that are as efficient as the bloodstream or neurons, so for them, almost all signaling is local and diffusion based. In addition, there are a number of examples of cells that release their own signaling molecules and then detect them with receptors. Here we look at the three types of short-range signaling in more detail. They can all be observed in Figure 07-02, below.
Paracrine signaling is considered to be a local signaling mechanism. The ligand is released into the extracellular space, and it diffuses through the extracellular matrix to be picked up by nearby receptors.
- As a result, cells that are farther from the source are expected to be exposed to a lower dose of the ligand. This kind of gradient-dependent ligand is heavily used throughout development, as different doses of the signal can be detected, and responded to, differently.
- Compared to endocrine and neuronal signaling, described above, the affinity for the ligand is likely to be more moderate. However, it will also be somewhat more variable than in other forms of signaling. Dose-dependent responses often use receptors with different affinities for the ligand so that they cannot be activated at concentrations that are lower than their sensitivity. Again, this is very common during embryonic development across kingdoms.
- Examples of paracrine signaling include the synaptic signaling we discussed earlier, where the neurotransmitter is released into the synapse and received by the receptors on the other cell in the synapse. Also, many of the growth hormones used in development will have dose-dependent responses, thus allowing multiple responses from the same growth hormone.
Autocrine signaling is when the signal is both released and received by the same cell. It is considered a type of paracrine signaling, since the signal must diffuse to the receptor through the extracellular environment.
- This form of signaling is often used by the immune system in order to help it ramp up the immune response when activated. It is also something that happens in cells that have become cancerous; it helps the cancerous cells break free of the normal regulatory controls so that they can grow and divide without restriction.
- Sometimes a signal is released by a cell and is received by both itself and neighboring cells. Like much in biology, and in signaling, these categories are not mutually exclusive, and a single ligand can be used in a number of ways to illicit cellular responses.
Finally, juxtacrine signaling isn’t very common overall compared to the other kinds of signaling. It’s also known as contact-dependent signaling, which gives you an idea of what is involved in this form of signaling. In this case, the cell that receives the signal comes into direct contact with the one that is sending out the signal. Most commonly, this would mean direct cell-to-cell contact, but it may also be an interaction between a cell-surface receptor and a glycoprotein of the extracellular matrix.
- Just because this form of signaling is not common, we don’t want you to get the impression that it’s not important when used. Some of the most important survival signals are juxtacrine. Integrins are receptors that bind directly to the extracellular matrix and help the cell know that it’s not (yet) time to undergo programmed cell death (also known as apoptosis). Complete loss of integrin binding, without undergoing programmed cell death, is also considered a hallmark of cancer and the start of the process known as metastasis, which is when cancerous cells migrate and form new tumors in different parts of the body.
- Another example of cell-to-extracellular matrix signaling is when leucocytes use this form of binding to “roll” through blood vessels (Video 07-02) prior to leaving the bloodstream at the site of an infection.
The most famous example of cell-to-cell juxtacrine signaling is the Notch-Delta signaling cascade, which is essential to embryonic development in animals. The receptor (Notch) is a plasma membrane protein on one cell, and the ligand (Delta) is on the other. When they bind, the cytosolic side of the Notch receptor is cut off and becomes a transcription factor that then enters the nucleus. As a result, the transcription factor activates gene expression, and the “fate” of this cell is permanently and irreversibly changed. Notch-Delta signaling is extremely important in development, when cells are gaining their identities.
The Stages of Cell Signaling
At the most basic level, signaling happens in the following stages:
- A signal is sent. There are a number of ways this could happen. A cell could release a molecular cue, or the environmental conditions themselves could provide the molecule that is being detected. We’ll do a more in-depth exploration of signals and ligands shortly.
- This signal is received by the cell. This will require a receptor to recognize the signal and respond to it. Most often the receptor is at the cell surface, since that’s where signals would arrive first, but it doesn’t have to be. Nitric oxide (NO) is an example of a signaling molecule that can diffuse across cell membranes, so its receptor is in the interior of the cell.
- The signal is “interpreted” by the cell that receives it. This could involve a number of things, like splitting the signal so that multiple responses are possible; it also likely involves transferring the signal across the plasma membrane by activating specific internal responses. This interpretation step is called transduction.
- Finally, the cell responds to the signal. A cell could have multiple responses to a signal. Some responses are fast, while others are slow. If the signal is used to start a process, like cell division, apoptosis, or changes in cell identity, then there could be multiple changes in the cell as it prepares for the new behavior.
We’ll look at each of these parts in turn as we continue to explore this topic.
1. Signaling Molecules Are Varied
Signaling events start with a signal that originates from the cell exterior. These signals could be something directly from the environment or something that originated from another cell (which may be part of the same multicellular organism or not). There are very few rules for what constitutes a signaling molecule other than the fact that there is a receptor capable of receiving the signal and responding to it. Some examples of signals include the following:
- Odorants: Each of the chemicals you taste and smell has a receptor that can bind to it and sends a slightly different signal to your brain so that you can tell the difference between them.
- Animals that are considered to have a better sense of smell will have many (thousands!) more receptors available than those that are not considered to have a good sense of smell. The increased number and variation in their receptors mean that their sense of smell is more nuanced comparatively. Among the vertebrates, mammals and fish are thought to have relatively good senses of smell, whereas birds are thought to have much worse senses of smell compared to other vertebrates.
- In the mammals, the grizzly bear is thought to have a truly spectacular sense of smell—possibly one of the best in the world. It is said to be 7 times stronger than that of a basset hound (the superstar sniffer of the dog world). In turn, basset hounds are thought to have a sense of smell that might be as much as 100 million times more sensitive than our human noses. Interestingly, but maybe unsurprisingly, whales have a terrible sense of smell compared to their fellow mammals. However, they have an excellent sense of taste.
- Biological macromolecules: There are examples of signaling molecules from all four of the biomolecules (lipid, protein, carbohydrate, nucleic acid). Insulin, which helps you regulate blood sugar, is a short peptide. Glucose itself acts as a signal for the release of insulin.
- Neurotransmitters: They’re usually small chemicals that are synthesized in the cell for eventual release. We’ve seen examples of these already (acetylcholine, dopamine, epinephrine, serotonin, etc.).
- Photons: There are many receptors that are able to respond to light. The photoreceptors in our eyes are a great example, but there are further examples across the various kingdoms of life. Photons are often used to help organisms regulate circadian rhythms, among other things, so that they know when it is time to sleep and when to be awake. Plants use light sensing extensively to decide when and how to grow and when to flower to maximize the change that their pollinators will be available.
- Mechanical force and other nonchemical signals: Your skin can respond to a variety of nontraditional signals, like physical pressure and temperature. Your ears respond to sound waves. Plants can also sense a number of the physical properties of their environment, such as temperature and gravity. They can even respond to injury (usually as a result of herbivory) and manage the wound while also ramping up their chemical defenses.
Most of the signals we’re going to explore in this chapter will be of the type that are released by one cell and then travel to the receptor in one of the ways described earlier. As such, they will most likely be small chemicals (like neurotransmitters) or biomolecules (like short peptides or carbohydrates).
2. Receptors Are Used to Receive Extracellular Signals
A cell’s first point of contact with the outside world is when one of its receptors binds to a ligand and a signaling pathway is activated. As mentioned above, since the ligand is coming from the exterior of the cell, it is quite common for the receptor to be on the cell surface, with the ligand-binding domain facing outward. However, if the ligand is able to diffuse across membranes (Aside: What chemical properties should the ligand possess to make this possible?), then the receptor could be somewhere inside the cell. We won’t be discussing too many examples of internal receptors, but the following are a few examples of signals that can cross the plasma membrane:
- Nitric oxide (NO) is detected by the nervous system and results in a relaxation of smooth muscle cells. NO is a very small, nonpolar molecule, so it slips easily through the plasma membrane without help. The receptors for NO are inside the cell.
- Estrogen, testosterone, and other sterol-based hormones are often received inside the cell by a family of receptors known as the nuclear receptors. Nuclear receptors are receptors that either live full time in the nucleus or move from the cytosol to the nucleus upon activation. They bind directly to the DNA and act as a transcription regulator when in their active state.
- Auxin is a key plant hormone that appears to be involved in…well…everything! It has a strong influence on growth and development. Since plants cannot get up and move from where they were planted, they respond to their environment, in part, by growing toward or away from a specific stimulus. Auxin helps with this kind of differential growth. It is also a small, uncharged molecule that is able to diffuse right into the nucleus. The auxin receptor is also a transcription factor, even though it’s not genetically related to the nuclear receptors described above.
- As you know, light is also a signal that is received and processed by many organisms and can penetrate the cell without help. While the light receptors in the rod and cone cells of the human eye are officially on the cell membrane, they are within a specialized structure where the plasma membrane has folded in on itself many times, so they’re no longer on the surface per se. In plants there are light receptors that are found it the cytosol, in addition to the photosynthetic apparatus, which is inside the chloroplast (as you know from Chapter 5).
A common theme for these internal receptors is that the activated pathway is often very short. In the case of auxin and the nuclear receptors, it is the receptor itself that enters the nucleus and causes the change in gene expression. From the perspective of exploring the principles of signaling, that doesn’t give us very much to work with. So in this chapter, we mostly focus on plasma membrane–bound receptors, as those tend to activate longer, more complex pathways; thus, they require more explanation to understand. A ligand that is received by a receptor on the plasma membrane will usually not enter the cell. Instead, the signal is transduced across the plasma membrane and then activates a series of events inside the cell that allow the cell to respond to the signal. All of these events together are known as the signaling cascade, and understanding how these cascades fit together is the purpose of the rest of this chapter.
Signaling cascades are often categorized by the type of receptor used to initiate them. These fall into several categories based on their structural features. Later in this chapter, we will explore some of the most common types of receptors and the responses that they activate.
3. Once the Signal Is Received, It Must Be “Interpreted” by the Cell
The binding of the ligand to the extracellular portion of the receptor causes a conformation change in the receptor that carries through the membrane and into part of the receptor that is inside the cell. This is how the signal “passes through” the membrane, and it’s the first step in a process called signal transduction. Transduction is when a signal is converted from one form to another. In this case, the extracellular signal molecule binds and initiates the conformation change in the receptor, which activates it and allows it to generate a response inside the cell.
Once the signal is perceived inside the cell, there are several things that could happen (Figure 07-03). However, not all possibilities will occur in every cell. Different cells express different genes, which will code for different signaling cascade components. Thus, a single ligand can have different effects in different cell types. This is helpful in multicellular organisms, as it means that an organismal-level response can be coordinated using a single signal molecule. An excellent example would be our response to adrenaline. When our body releases adrenaline, there are a wide variety of physiological effects (e.g., increased heart rate and breathing, perspiration) that are all designed to prepare us for the “fight or flight” response to danger. Each of these physiological responses begins when a receptor on different cell types binds to adrenaline, and a signaling cascade is triggered.
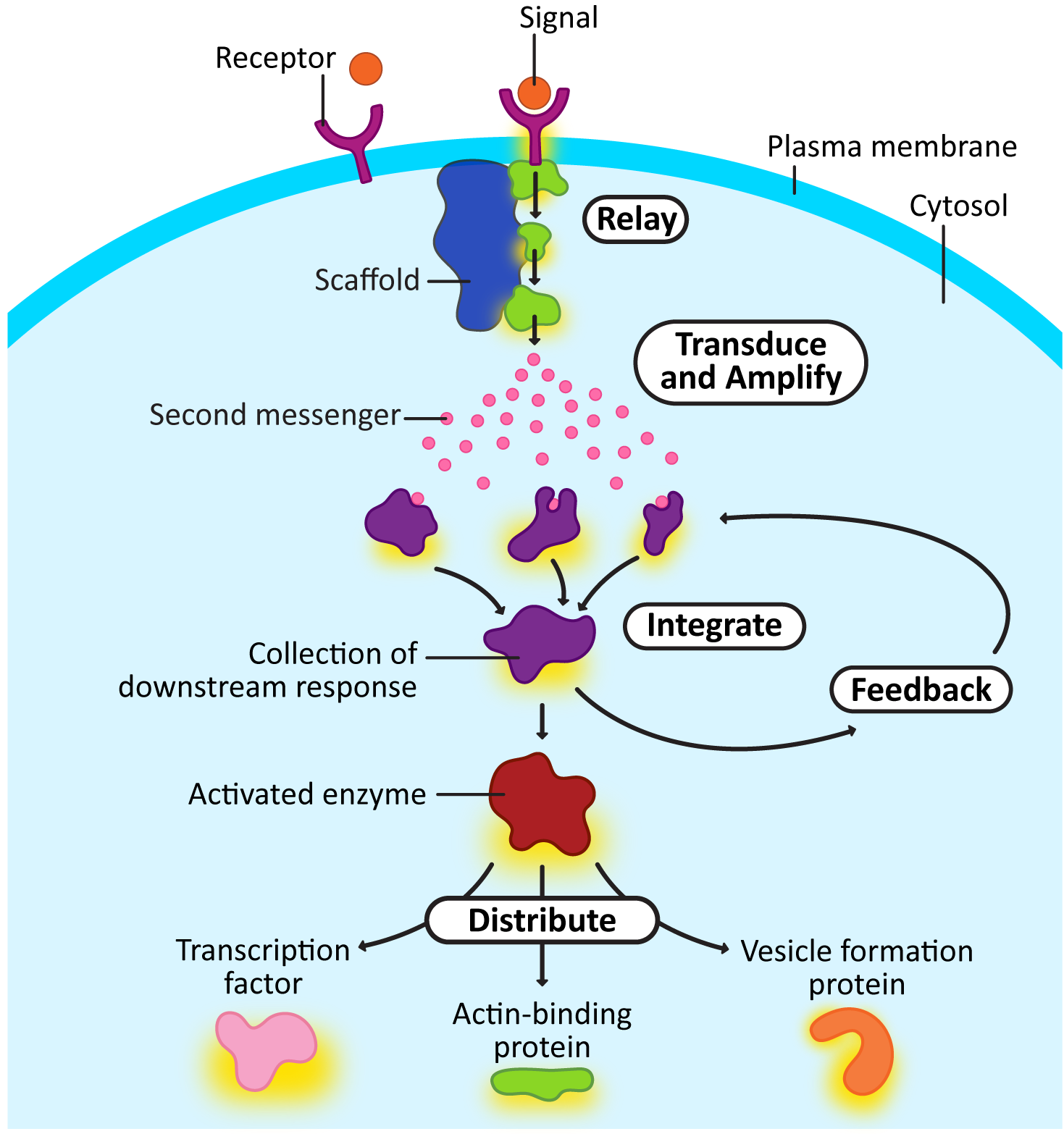
Here’s a quick explanation of each of the intracellular signaling steps from Figure 07-03. Later, we’ll look at some of the more famous examples of proteins that perform these roles. Remember that not all of these are present in every signaling cascade, and sometimes an individual element could be performing more than one function.
- Relay: Relay is an event where the signal is passed from one intracellular signaling molecule to the next. This is not dissimilar from passing a baton in a relay race. In Figure 07-03, this is shown right after the receptor, but in reality, it can happen anywhere in the signaling cascade. Often, the “signal” that is being passed along is the addition or removal of a phosphate group on a target protein (known as phosphorylation or dephosphorylation, respectively). You’ll also notice a larger protein that connects three intracellular relay proteins together. This protein is called a scaffold protein. They often aid in relay signaling events. Their job is to hold several proteins together so that activation can be more efficient.
- Transduce and amplify: Often, a signal needs to be transformed so that it can be used in other ways. Phosphorylation is a common modification in signaling cascades but only works on other proteins. Small “messenger” molecules (often referred to as second messengers) are used to amplify the signal and quickly spread it throughout the cell. These molecules are either produced by proteins that have been activated as part of the response or released into the cytosol from another compartment (like the ER). Some of the most famous second messengers include the following:
- Calcium ions flood the cytosol after they have been released from the ER.
- Cyclic AMP (cAMP) is produced quickly in the cytosol from adenosine monophosphate (AMP).
- PIP2, a specific phospholipid molecule, can be split into a membrane-bound second messenger called DAG (diacylglycerol) and a soluble second messenger called IP3 (inositol triphosphate).
- Integrate: In this process, multiple receptors feed into a single “downstream” response. In some cases, both signals will be required for the response to occur, while in other cases, either signal can cause the response in question, regardless of whether the other is present.
- Feedback: As the signal moves through the cascade of responses, later events can influence earlier components of the pathway. We call this a feedback loop. Feedback loops can be either positive (i.e., later events help further activate earlier steps) or negative (i.e., later events stop earlier events from continuing).
- Distribute: At some point, it’s probable that an enzyme will be activated whose function is to activate many different proteins. The proteins that are being acted upon will have a wide variety of functions (e.g., transcription factors, actin- or microtubule-binding proteins, vesicle-formation machinery). This is how the signal is distributed so that cellular function changes.
4. Responses to the Signal Can Also Be “Fast” or “Slow” and Are Usually Both
The cellular responses at the end of the signaling cascade usually fall into two categories: fast responses and slow responses. The difference between these is very simple. Cellular responses that require protein synthesis will always be slower than responses that rely on preexisting proteins, which can be simply altered.
Slow Responses Come from Changes in Gene Expression
Often, a signaling event will result in an overall change in cellular identity. Changing the identity of a cell will require a number of changes in gene expression: some proteins will need to be synthesized, while others will likely need to be removed from the cell. For example, most of the signaling events involved in development will cause changes in gene expression, as cell differentiation is a key step in embryonic development. As a secondary example, the immune system also uses alterations in gene expression so that antibodies can be produced in response to a pathogen or vaccine. In order to change gene expression, signaling cascades cause the activation or deactivation of transcription factors or proteins that affect chromatin structure (i.e., histone or chromatin-modifying enzymes). As we discussed in Chapter 3, proteins that influence histones and/or chromatin will impact the accessibility of the DNA, thus impacting the expression of genes in that area. The end result will be changes in the protein composition in the affected cell.
Fast Responses Come from Changes in Preexisting Proteins
Alternatively, extracellular signals can stimulate signaling cascades that result in shorter-term changes in cellular function by altering preexisting proteins. A very good example of this, which we will explore in detail in Chapter 8, is mitosis. When mitosis is activated, the cell is vulnerable, so this process must be completed quickly. Most of the proteins involved in rearranging the cytosol for mitosis already exist before mitosis starts. They are present in the cytosol, waiting for the signal to initiate cell division. Once the signal is perceived, these preexisting proteins are modified as a result of the signaling cascade. This causes a rapid shift in cellular physiology, and the cells progress through mitosis as efficiently as possible.
Deactivation of a Signaling Cascade
So far in this chapter, we have discussed how signaling cascades are activated. In some cases, this is the end of the story, as the signal never needs to be turned off. An example of this would be the Notch-Delta signaling pathway, which changes the cell permanently. It doesn’t really need to be “turned off” in the traditional sense. Another great example would be apoptosis. The signaling cascades of apoptosis lead directly to the death of the cell, so once apoptosis is activated, there is no cellular mechanism to deactivate it.
On the other hand, a great many signaling events need to stop eventually so that the cell can go back to “normal.” Mitosis is a good example, as the signaling that started mitosis must be stopped in order for mitosis to end. Another good example is sensory perception. Every single one of our senses is controlled through signaling. If we can’t stop receiving and responding to the signal, then we can’t go back to normal so that the signal can be received again. Imagine if our pain receptors never turned off once they were turned on!
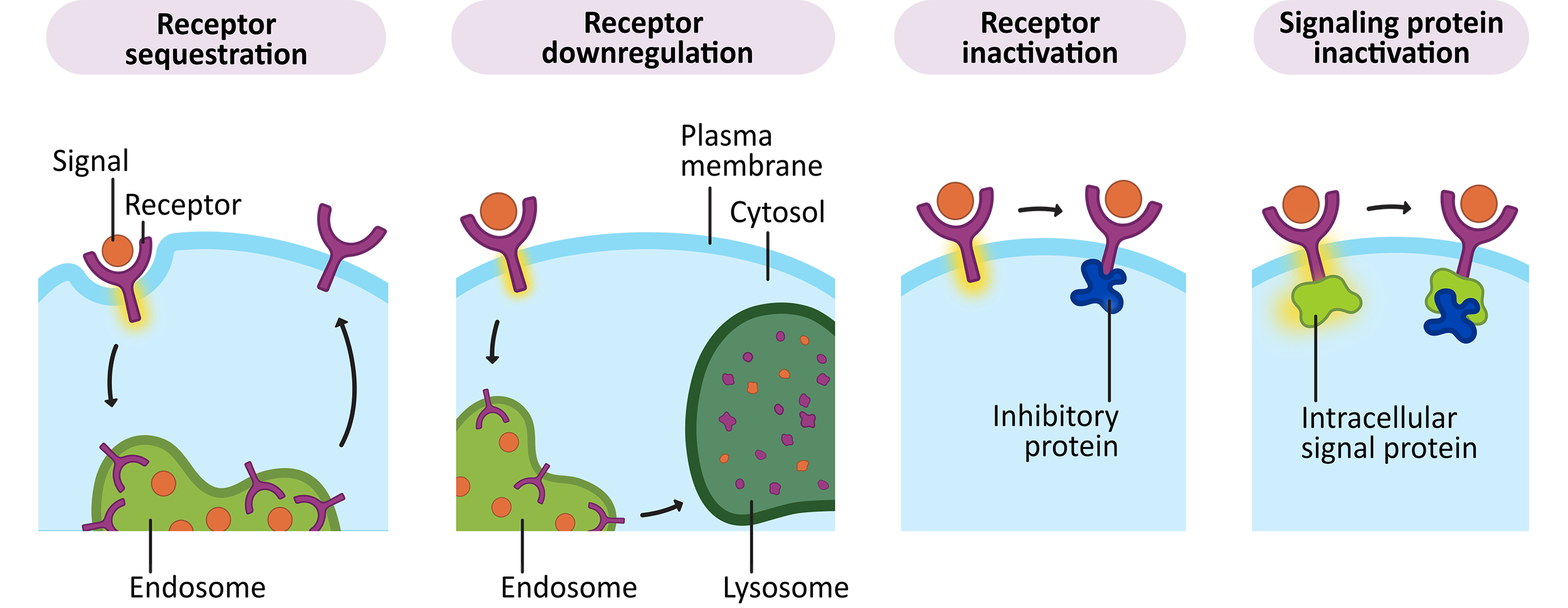
Figure 07-04 illustrates the most common ways that signaling cascades are stopped. Quite often, cells deactivate signals by inhibiting, or removing, the receptor. In some cases, it is an internal component that is deactivated.
Some of these changes are permanent. An example of permanent deactivation occurs in receptor downregulation, where the receptor is brought into the cell via endocytosis and then degraded in the lysosome. However, many other deactivation steps are reversible. Receptor sequestration pulls the receptor off the cell surface so that it can’t bind to a signal for a period of time but the receptor isn’t destroyed. In both receptor inactivation and signaling protein inactivation, one of the proteins that forms part of the signaling cascade is inactivated in some reversible way (most likely by phosphorylation and/or binding to an inhibitory protein).
Deactivation of signaling cascades is usually controlled via negative feedback loops. This means that the activation of the signaling cascade produces products that help shut down the signaling cascade as well.
Studying Cells: Reading and Interpreting Diagrams of Signaling Cascades
Before we move on to the next topic, we should take a moment to discuss the way that scientists write out signaling cascades and how to interpret them.
As you likely have seen in the figures here or elsewhere in your course notes or the internet, signaling cascades are often written out with the proteins lined up and connecting symbols between them. Two important symbols are used in between the proteins to identify how they impact each other:
- a regular arrow ( → ), which is used when protein 1 activates protein 2, and
- a blunt arrow ( ⊣ ) , which is used when protein 1 inhibits the function of protein 2.
We often discuss the steps of a signaling cascade as being upstream or downstream of each other. So for example, the signal binding to the receptor is upstream of the cellular response. Conversely, the response will be downstream of ligand binding. These terms and symbols are important for interpreting the diagrams used to represent signaling pathways in textbooks and papers. Video 07-03 provides further explanation.
Topic 7.2: Examples of Common Signaling Mechanisms
Learning Goals
- Compare and contrast ion-gated channels, G-protein-coupled receptors (GPCRs), and enzyme-coupled receptors, including their mechanism of activation, response, and deactivation.
- Name and describe the possible downstream pathways used after GPCR and enzyme-couple receptor activation. Identify which ones are more commonly used with each type of receptor.
- Explain how researchers can experimentally investigate the role of posttranslational modifications in the proper functioning of a cell signaling cascade.
All right! Now that we have a better understanding of some of the general principles of signaling, the next step is to look at examples of the proteins that do this work and also how they do it. As you may have already noticed, signaling can be quite confusing. There are a lot of components that interact, so it can be difficult to keep it all straight. Our suggestion is to take it slow and to stop and think about each step and how it might influence the others. Memorizing will not be very helpful here, as there are too many things to keep track of, so it’s better to take a step back and try to understand what’s going on. We’re not going to dive too deep, since this is still only a brief introduction to the beautiful complexity of signaling.
We’ll start by discussing the most common cell-surface receptors (Figure 07-05 and Video 07-04). Along with each receptor type, we’ll also examine some of the more common “downstream” responses to each receptor type. Remember that there is quite a lot of mixing and matching when it comes to the proteins involved in signaling cascades.
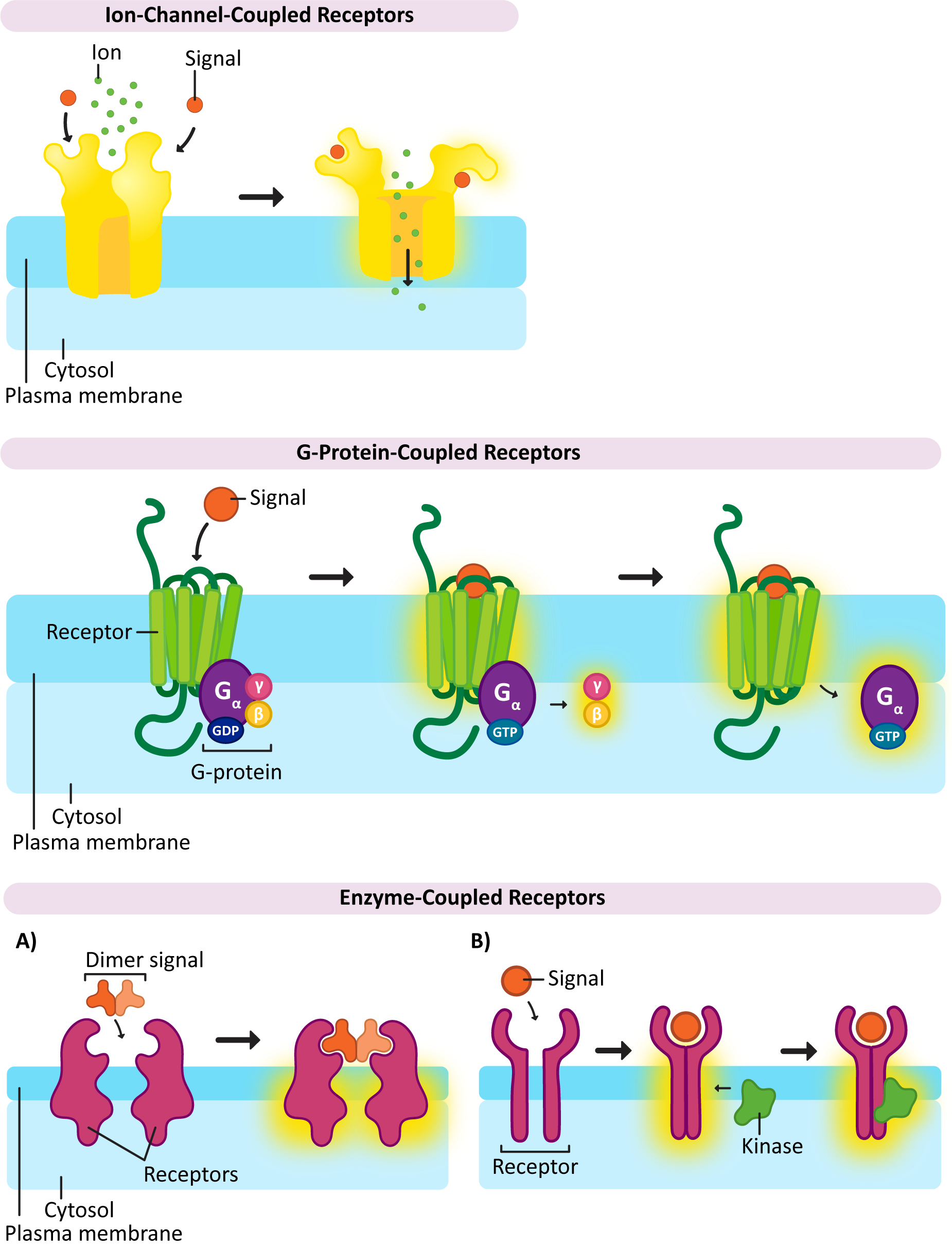
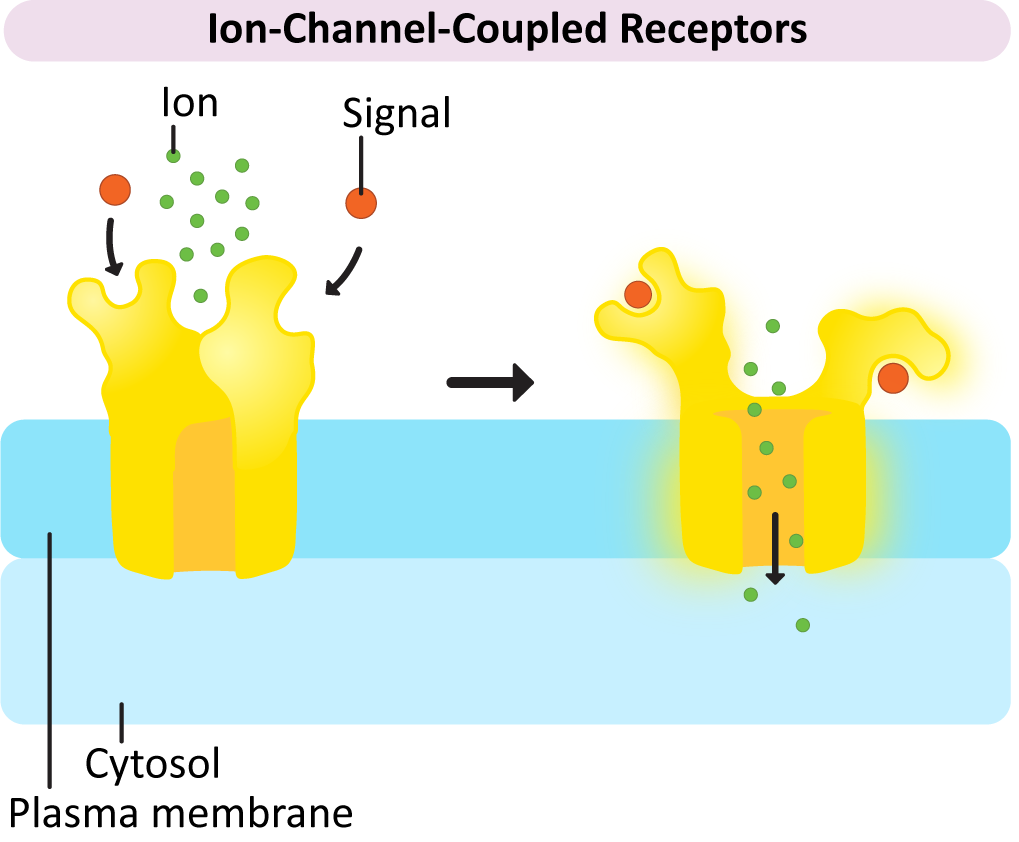
Ion-Channel-Coupled Receptors
As the name says, ion-channel-coupled receptors are receptors that, when activated, open (or close) an ion channel (Figure 07-06). This is very common in the nervous system, where action potentials are produced through ion channels and perpetuated in part by the release of neurotransmitters. In the case of the nervous system, the purpose of these ion-channel-coupled receptors is to convert a chemical signal (like a ligand) into an electrical one. Unfortunately, this aspect of neuronal function is well beyond the scope of this textbook. As a result, this function of ion-channel-coupled receptors is not going to be discussed much. Another name for an ion-channel-coupled receptor is a ligand-gated ion channel.
There is one important example of a ligand-gated ion channel that we will be discussing a bit later, and that’s the one used to release calcium into the cytosol from the ER. Remember that calcium is an important second messenger in cell signaling and is often used to amplify and spread the response throughout the cell. The ER-localized calcium channels that are used in this process open to a specific signal as well. In this case, another second messenger is used, IP3, which is released when the phospholipid, PIP2, is cleaved as part of a signaling response. We will see how this works a bit later in this topic.
G-Protein-Coupled Receptors (GPCRs)
The next group of receptors we’ll look at is G-protein-coupled receptors, or GPCRs for short. This type of receptor is by far the most common in animals. They are heavily involved in animal sensory perception, especially our sense of smell. Humans have over 700 GPCRs in total, most of which are dedicated to smell. On the other hand, a mouse, which has a much sharper sense of smell than we do, has over 1,000 different GPCRs involved in its sense of smell alone!! Even better sniffers, like the basset hound and grizzly bear we talked about earlier, are thought to have an even higher number of olfactory GPCRs.
GPCRs are a very large family of proteins that includes many subfamilies with specific functional differences. We won’t get into all of that detail here (it fits better with an upper-level signaling textbook), but we can identify some important characteristics of GPCRs that are common across the family. For example, the GPCR itself is almost always a single polypeptide chain with seven transmembrane domains that cross the membrane as alpha helices. This is considered to be a characteristic structural feature of these receptors. GPCRs have a large pocket for ligand binding, which allows for a lot of variation. Thus, the ligands that can bind to GPCRs are also quite variable. This is why they are such a good candidate to manage our sense of smell—they most commonly bind to small organic molecules rather than peptide hormones. Some of the light receptors on our retinas are GPCRs as well.
Once again, the name of this type of receptor tells you something about it. The receptor itself works in conjunction with another set of proteins called G-proteins. The “G” in G-protein stands for GTP, as GTP is required to activate the G-proteins and allows a response to be generated. G-proteins also come in a variety of flavors to match up with the GPCRs that they work with.
Despite variation in the G-protein family, they also have many similarities:
- Each G-protein complex contains three subunits: the alpha (α), beta (β), and gamma (γ) subunits.
- The three subunits can all bind together in a trimeric complex, or G-alpha can separate from the other two. G-beta and G-gamma almost always stay together as a unit.
- The alpha-subunit is able to bind to GDP or GTP. Both the alpha- and the gamma-subunits have covalently linked lipid tails, which allow them to remain associated with the membrane.
- The beta unit does not have a lipid tail, but since it remains attached to the gamma-subunit, it also remains attached to the cell membrane.
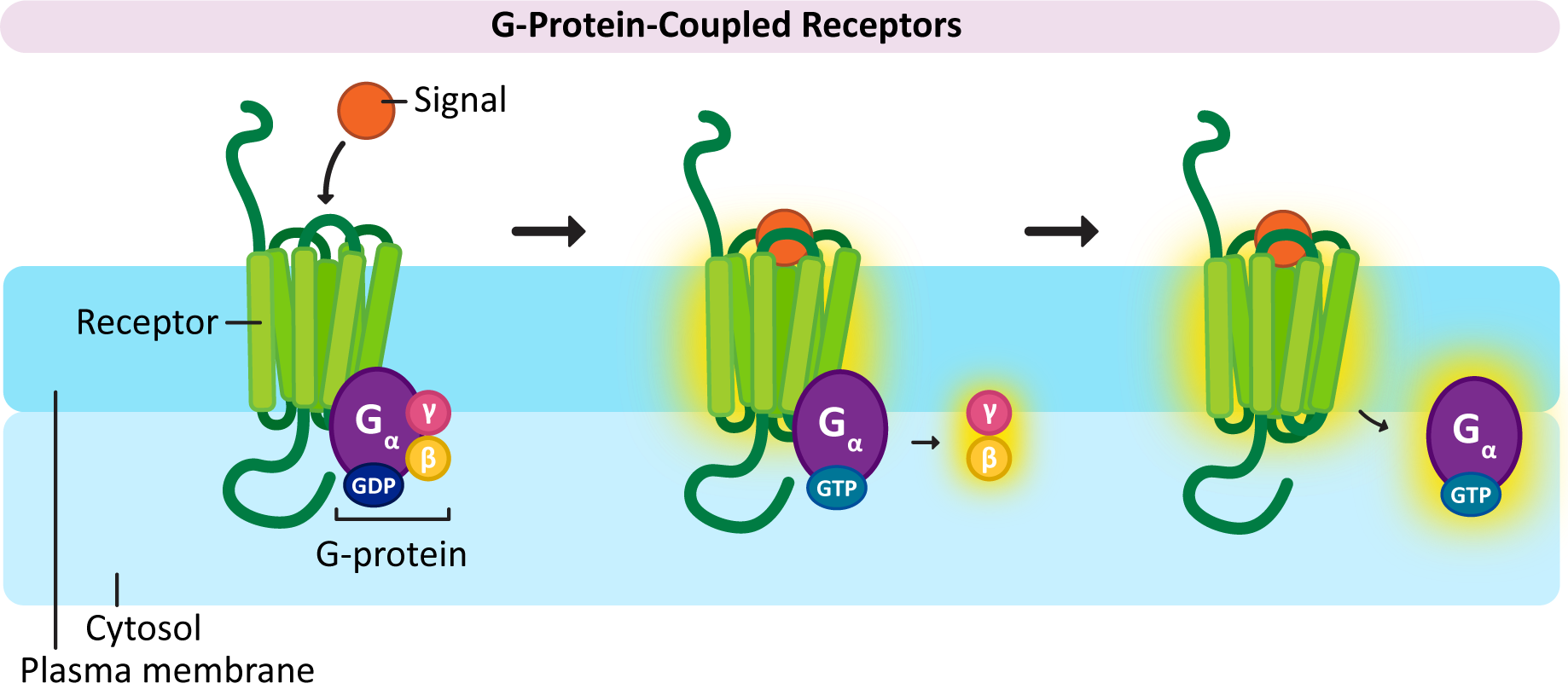
In a nutshell, here’s how GPCR activation works (Figure 07-07):
- When the receptor binds to its ligand, it undergoes a conformation change and binds to the α/β/γ complex. (At this point, the three subunits are bound together as a single complex attached to the GPCR on the cytosolic face of the plasma membrane.) This causes the GDP to be released from the α-subunit and get replaced by GTP.
- The binding of the GTP molecule causes a conformation change in the G-protein complex, and the β/γ subunits are released from Gα.
- The separated G-protein subunits (Gα and Gβγ) are now considered active and activate their appropriate downstream targets. So by this mechanism, we effectively split the signal as it is being relayed so that we can create more change in the cell.
It’s worthwhile to note that as the G-proteins are activated and move away from the receptor, the lipid tails keep them associated with the interior surface of the plasma membrane. As we learned back in Chapter 2, membranes are fluid, so the G-proteins will be able to diffuse laterally through the membrane until they come into contact with proteins that they can bind to and activate. - Eventually the G-proteins will deactivate themselves. The Gα subunit will slowly hydrolyze the GTP so that it is converted back into GDP (and a phosphate molecule, which is released). This inactivates the Gα subunit, which causes all three G-proteins to return to their inactive state.
Because the activation of the G-proteins is dependent on the presence of GTP in the Gα subunit, this provides a built-in system for shutting down the response. The GTP is short lived, so the response lasts only as long as the GTP does. However, there may be many cycles of activation and deactivation of G-proteins during a single response.
Common Downstream Effects of GPCRs—Second Messengers: Cyclic AMP (cAMP)
Now that we’ve discussed how the GPCR is activated and deactivated, we turn our discussion to an important downstream signaling molecule commonly used in GPCR signaling cascades: the production/release of second messenger molecules. We’ll specifically discuss the two most common second messengers that are activated by G-proteins: cyclic AMP and IP3/DAG.
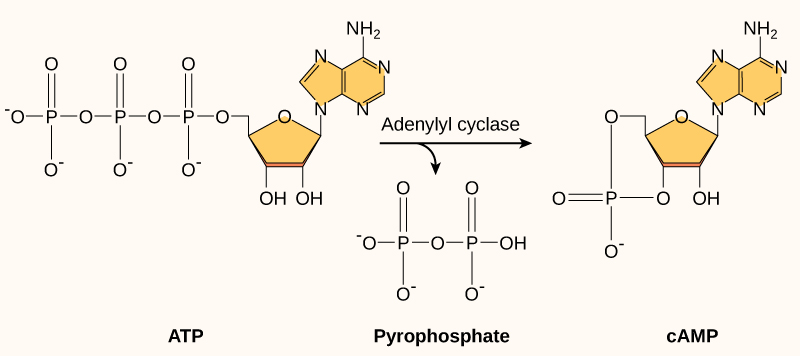
Cyclic AMP, or cAMP, as it is often abbreviated, is a modified version of adenosine monophosphate (AMP). cAMP is made from ATP, a molecule you should know well that is extremely abundant in the cell. Figure 07-08 shows the structural changes required to interconvert ATP, cAMP, and AMP.
Cyclic AMP is produced when a plasma membrane–bound enzyme, adenylyl cyclase, is activated by the Gα subunit of the activated G-protein (Figure 07-09). Cyclic AMP is then capable of moving throughout the cell and interacting with any enzymes that have binding sites for it. cAMP is a short-lived molecule—its half-life in the cell is less than 1.5 minutes. An enzyme known as phosphodiesterase breaks it down so that the cellular response is kept short. Figure 07-09 and Video 07-05 (below) review the activation of GPCRs and discuss the production of cAMP.
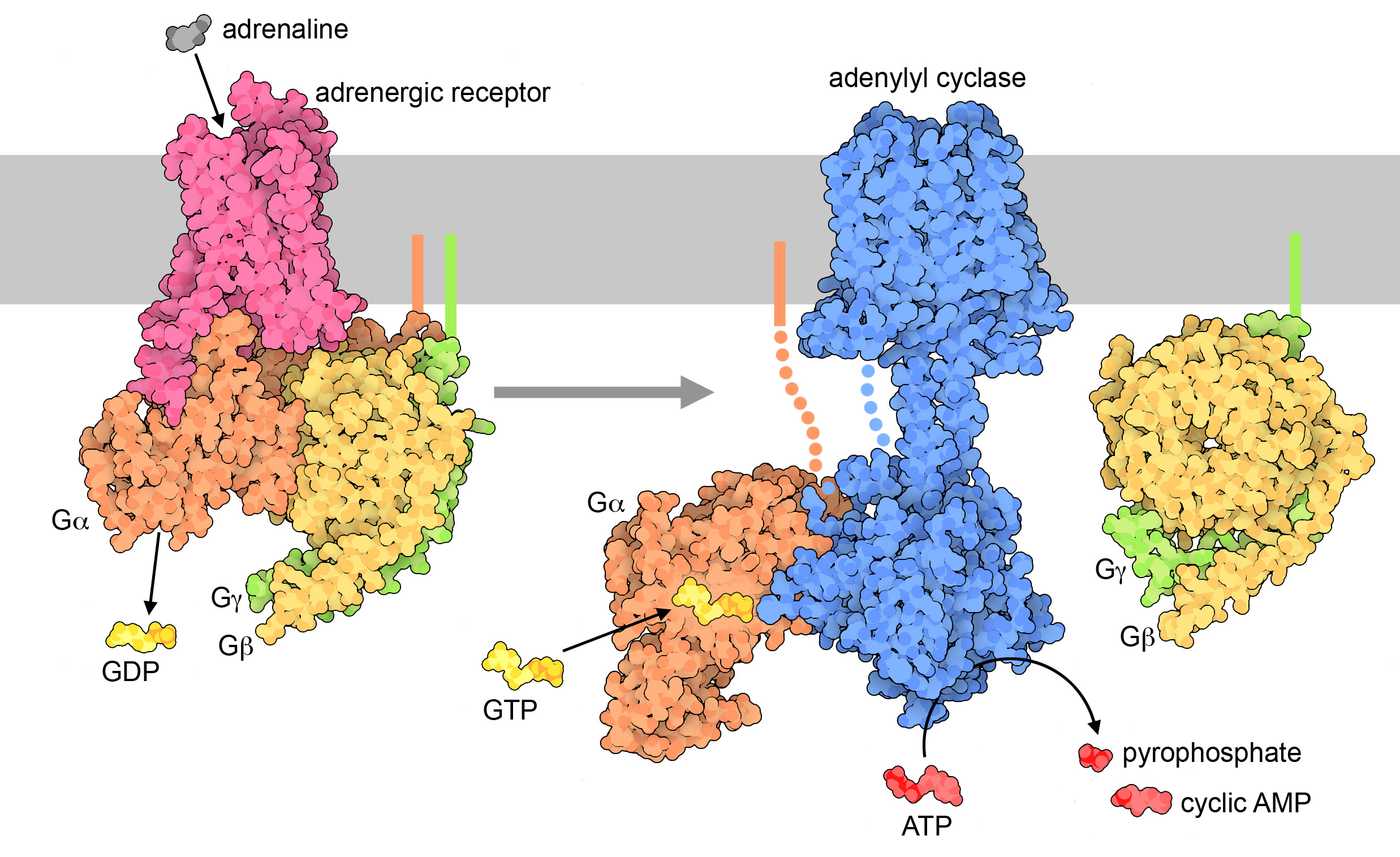
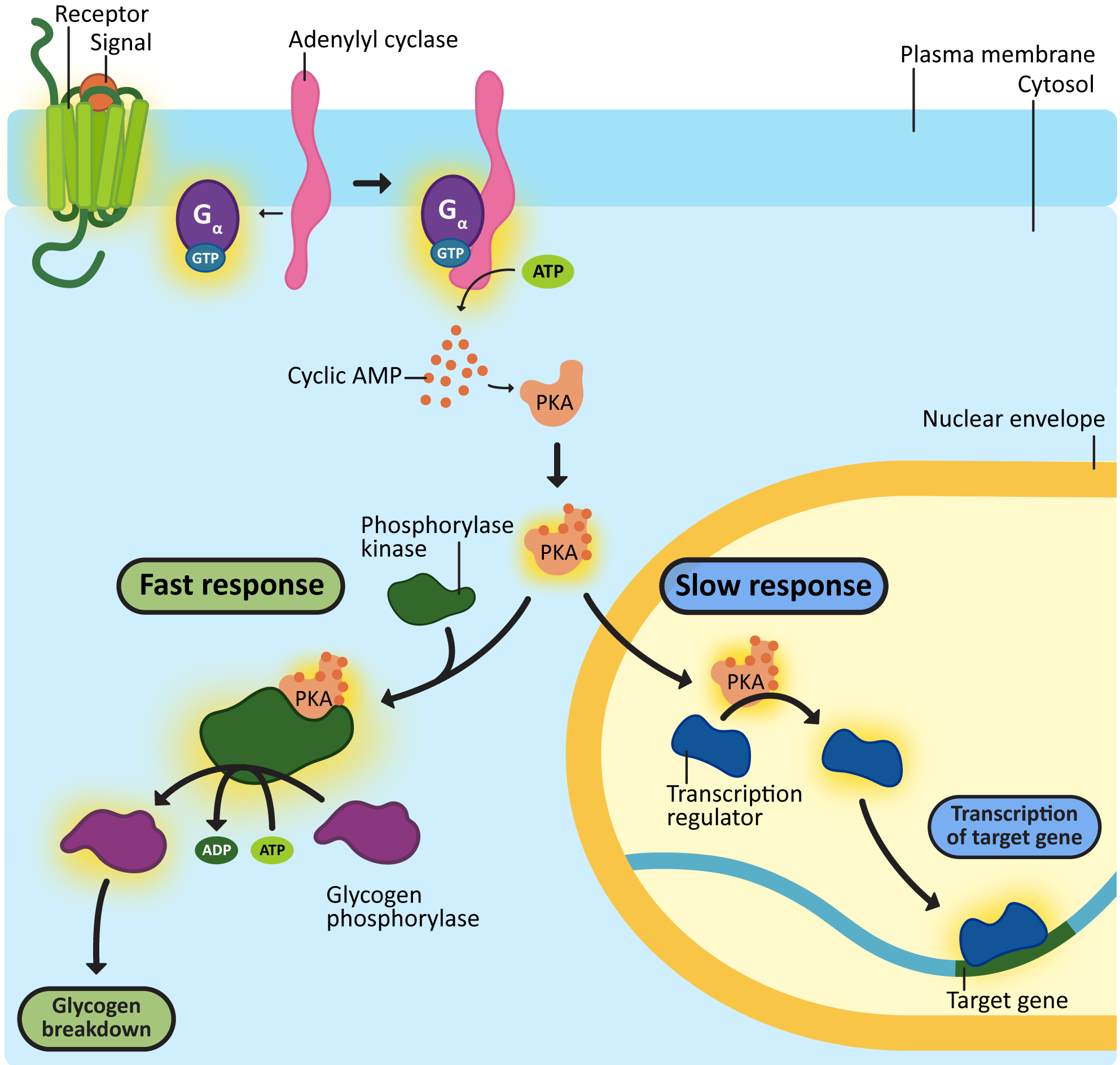
One of the most important enzymes activated by cAMP is a cytosolic protein called protein kinase A (PKA). When activated, PKA can have a number of different effects. Figure 07-10 shows two examples, one of which is a “fast” response, illustrated by the breakdown of glycogen in the liver. The other is a “slow” response—PKA can also enter the nucleus and phosphorylate transcription factors, which will activate these transcription factors, resulting in gene expression. Remember that different cells/tissues will have different proteins present in their cytosol that can be phosphorylated by PKA, which, in turn, will result in different effects.
Common Downstream Effects of GPCRs—Second Messengers: DAG, IP3, and Calcium
DAG and IP3 are another set of common second messengers. They are always produced together, as they are two parts of the same initial phospholipid (called PIP2, which is a generic term for a phosphatidyl inositol phosphate with two phosphates added to the inositol sugar). PIP2 gets split in two by an enzyme known as phospholipase C (PLC) Figure 07-11) to produce the two second messengers. PLC is activated by a Gβγ protein complex. While the breadth of lipid function in cells doesn’t often get discussed at the introductory level, it is worth noting that there are a number of membrane lipids, especially phospholipids, that have a key role to play in cell signaling and membrane identity.
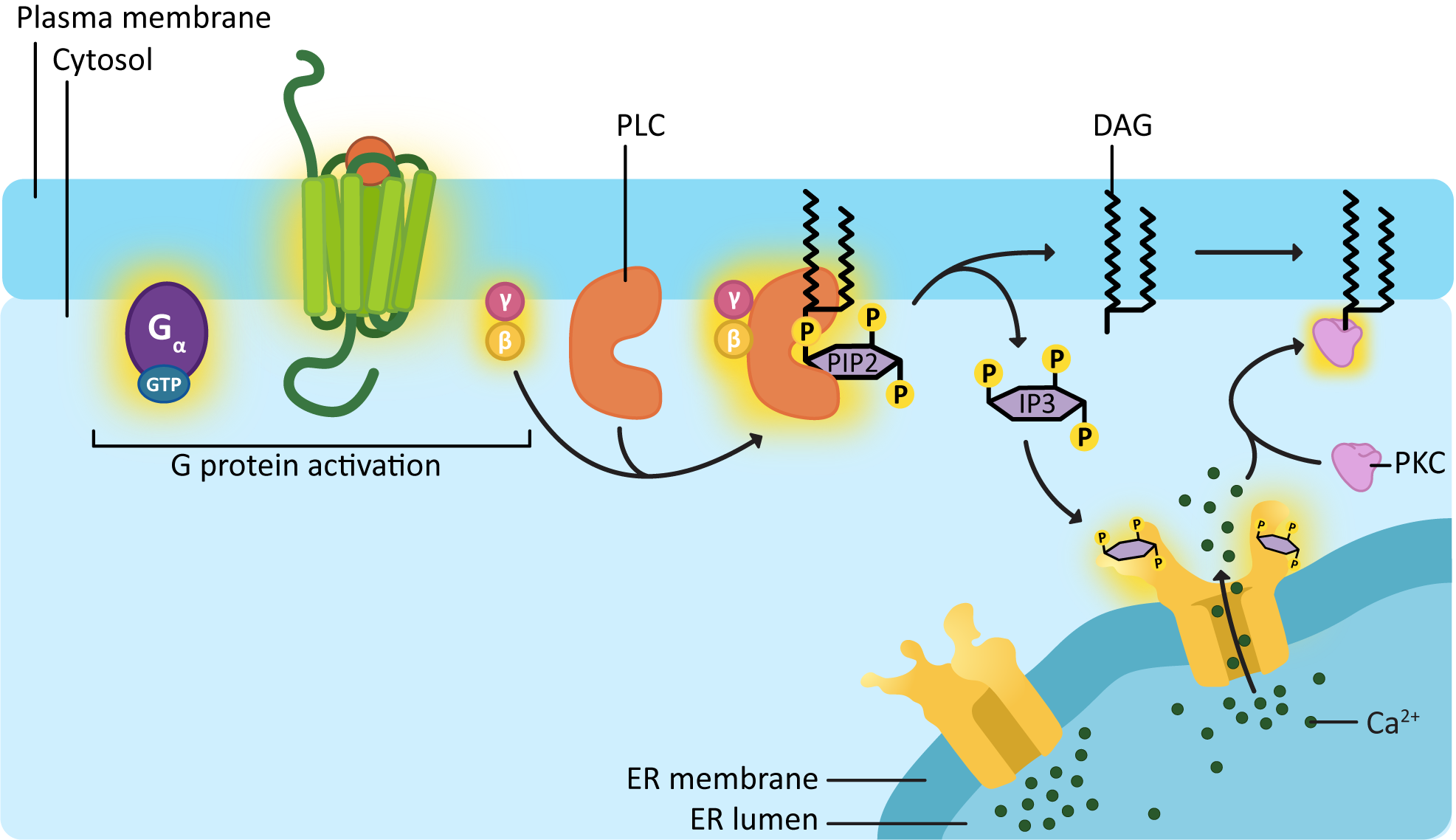
As illustrated in Figure 07-11, phospholipase C clips the soluble “head” portion of the phospholipid from the glycerol backbone and lipid tails. The head portion is a specific type of six-carbon sugar, with three phosphate groups on it, called inositol 1,4,5-triphosphate, or IP3 for short. The hydrophobic “tail” portion that is left in the membrane is now a molecule known as diacylglycerol, or DAG. Both of these molecules will be used to activate further components of the signaling cascade. DAG will diffuse laterally through the membrane and act as a docking site for a protein known as protein kinase C (PKC). IP3 is now soluble, so it can diffuse through the cytosol and bind to and activate its downstream targets.
One important protein that IP3 can bind to is a ligand-gated ion channel in the ER. This ligand-gated channel is responsible for releasing calcium ions, another important second messenger in the cell, into the cytosol. Much like cAMP, calcium can flood the cytosol very quickly, producing a rapid change in the membrane potential of the plasma membrane, among other things. Calcium waves that rapidly spread through the cell are a key event in many cellular processes, such as muscle contraction and oocyte fertilization.
The released calcium can also interact with many different targets in the cytosol. In Figure 07-11, we see that PKC requires calcium to bind in order for it to become active. So PKC activation requires both the docking site, produced by DAG, and the calcium released by IP3 before it can be activated. Activated PKC will then be able to phosphorylate a number of specific targets to generate change in the cell.
Eventually, the calcium is pumped back into the ER using different ion channels than those that were opened during calcium release. The new channels are opened by the presence of calcium in the cytosol, which makes this an excellent example of a negative feedback loop, where the signaling event activates the process that will also deactivate the signal.
Incidentally, calcium is by far the most commonly used second messenger in plants. Also of interest is that GPCRs are not nearly as common in plants as they are in animals. The most commonly used receptors in plants are receptors coupled to serine/threonine kinases. Ser/Thr kinases are enzymes, so these are a type of enzyme-coupled receptor. This leads us very nicely to the last type of receptor we need to discuss.
Enzyme-Coupled Receptors
This final set of receptors that we need to discuss is much more structurally variable compared to GPCRs, but they all have one very important thing in common: when the ligand binds to the receptor, an enzyme known as a kinase is activated. Kinases add phosphate groups to other proteins. We’ve seen many examples of this kind of enzyme throughout this textbook (specific examples include PKA and PKC from the previous section). There are also enzymes that remove phosphate groups, which we call phosphatases.
Phosphate groups can only be added to proteins at specific sites—not all R groups on an amino acid are capable of having phosphate groups added. Kinases fall into two major categories based on the amino acid R groups they modify:
- Tyrosine kinases: These are the largest class of kinases that associate with receptors and thus are better studied overall within the context of signaling. In this case, phosphates are added to tyrosine residues on the target protein. Tyr kinases are often also receptors, or directly associated with receptors, so these are the specific type of kinase we discuss most often in this section.
- Serine/threonine kinases: Serine and threonine amino acids have very similar sidechains in that they are short hydrocarbon chains with a reactive hydroxyl group (-OH). Thus, the kinases that work on them are structurally similar as well. As such, we call this group serine/threonine (Ser/Thr) kinases. These kinases are more common in plants and in soluble kinase proteins not associated with receptors. PKA and PKC, from the previous section, are Ser/Thr kinases.
The enzyme-coupled receptors themselves will either
- have their own intrinsic phosphorylation capacity, meaning that they can act on themselves or their dimerized neighbor, which is activated upon ligand binding, or
- have no intrinsic enzymatic activity but will be closely associated with a kinase that is activated upon ligand binding.
We won’t really differentiate between the types (integrated kinase or associated kinase) here, as there really isn’t a whole lot of difference between them from a functional standpoint. Additionally, for the most part, the examples we’ll see will be receptor tyrosine kinases (RTKs for short), where the kinase acts on tyrosine and is an intrinsic function of the activated receptor. Remember that there are also receptor-associated Ser/Thr kinases. Ligands for these kinds of receptors are often small peptides, such as growth hormones and insulin.
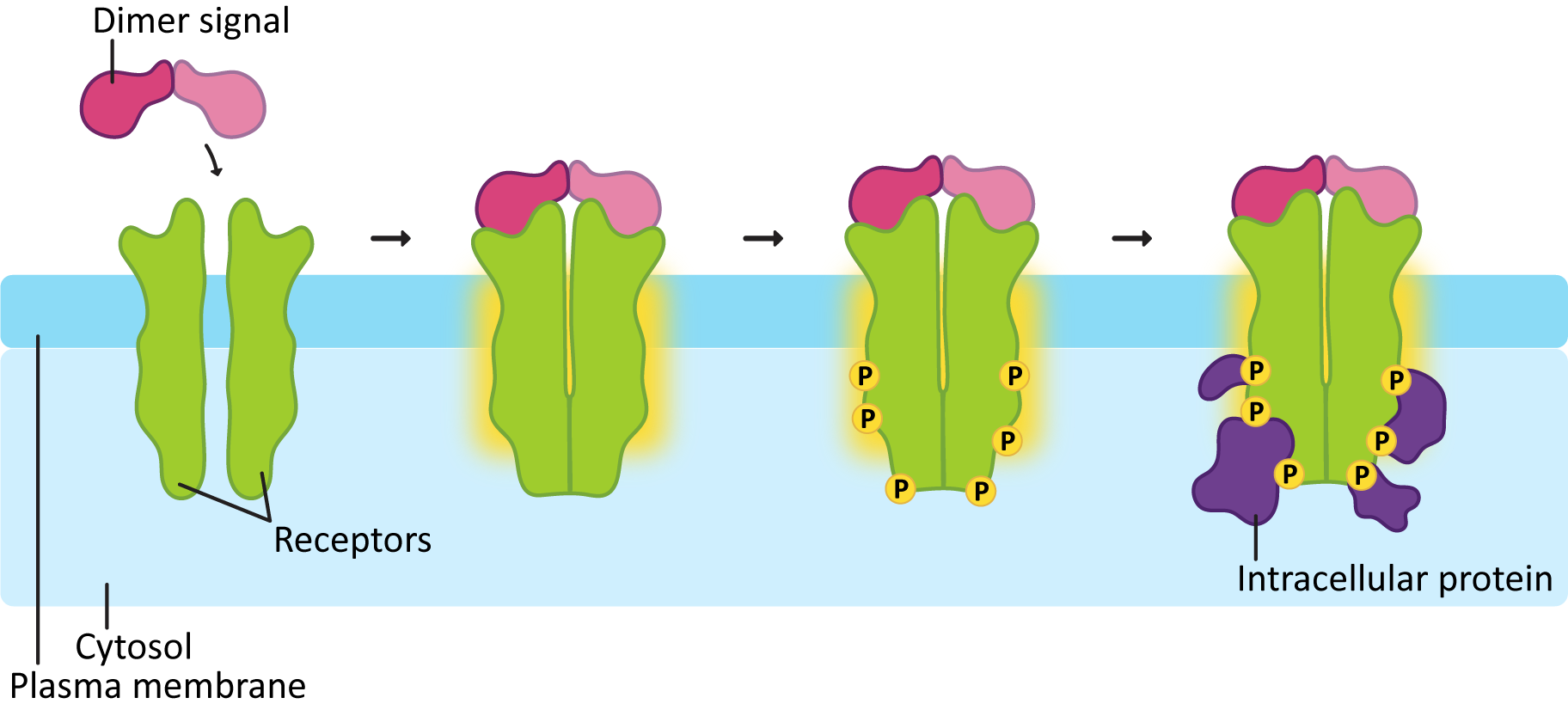
A few important points about RTKs are highlighted by Figure 07-12:
- The first is that RTKs usually dimerize as they bind to the signal molecule. So the inactive receptor is solo, and when the ligand binds, the receptor forms a dimer with either a second version of itself (i.e., a homodimer) or a closely related, but not identical, version (i.e., a heterodimer).
- Once they dimerize and the ligand is bound, the enzymatic activity is turned on. As a general rule, each protein in the dimer will phosphorylate the other one.
- The added phosphate groups on the receptor most commonly form the basis for docking sites so that a bunch of other proteins can bind and be phosphorylated. These docked proteins will help relay the signal to the next step. At this point the receptor is acting as a scaffold protein in addition to its role in phosphorylating target proteins.
Much like GPCRs, there are both membrane-bound and soluble proteins that will be activated by enzyme-coupled receptors. Second messengers may or may not be involved with these receptors, though we are not going to explore any examples of second messenger activation by RTKs here (we already did that). We’re going to look at two new examples of common downstream effects: Ras and mitogen-activated protein (MAP) kinase signaling as well as another example of lipids that have roles in signaling.
Downstream Effects of Enzyme-Coupled Receptors—Ras and MAP Kinase Cascades
Just like GPCRs, an activated receptor will need to relay the signal and amplify it somehow so that it can be spread around the cell and induce changes in the cell. We’ve already seen a couple of ways this can happen with GPCRs. In the case of RTKs, the first commonly activated molecule after receptor activation is a protein called Ras.
Ras is a molecular switch that is somewhat related to G-proteins. Molecular switches like these are very common in the cell. In fact, there is a very large family of these proteins (so big that we call it a superfamily) that is named after Ras. The members of the Ras superfamily of proteins are all small monomers (i.e., they are monomeric) and are all GTPases. This means that they convert GTP to GDP. We’ve seen many examples of GTPases in other chapters of this textbook, including the following:
- Ran, which helps proteins to enter/exit the nucleus (Chapter 3)
- Rabs, which help with vesicle docking (Chapter 4)
- GTPases, which are part of the vesicle coat (such as Arf and Sar) and help the coat fall off after the vesicles form (Chapter 4)
- Tubulin, a very distant relative of Ras that works in a very similar way (Chapter 6)
The reason that we call these proteins molecular switches is that they have an “on” and “off” conformation, which is determined by the presence of GDP (off) or GTP (on). This is extremely similar to the G-proteins we saw earlier when discussing GPCRs. Like the G-proteins, Ras also has a small lipid tail covalently linked to it, which is exposed when the protein is activated. Thus, when the receptor activates Ras, it will act very similarly to a G-protein in that it will associate with the plasma membrane, diffuse laterally, and activate other proteins at the cell surface, as shown in Figure 07-13.
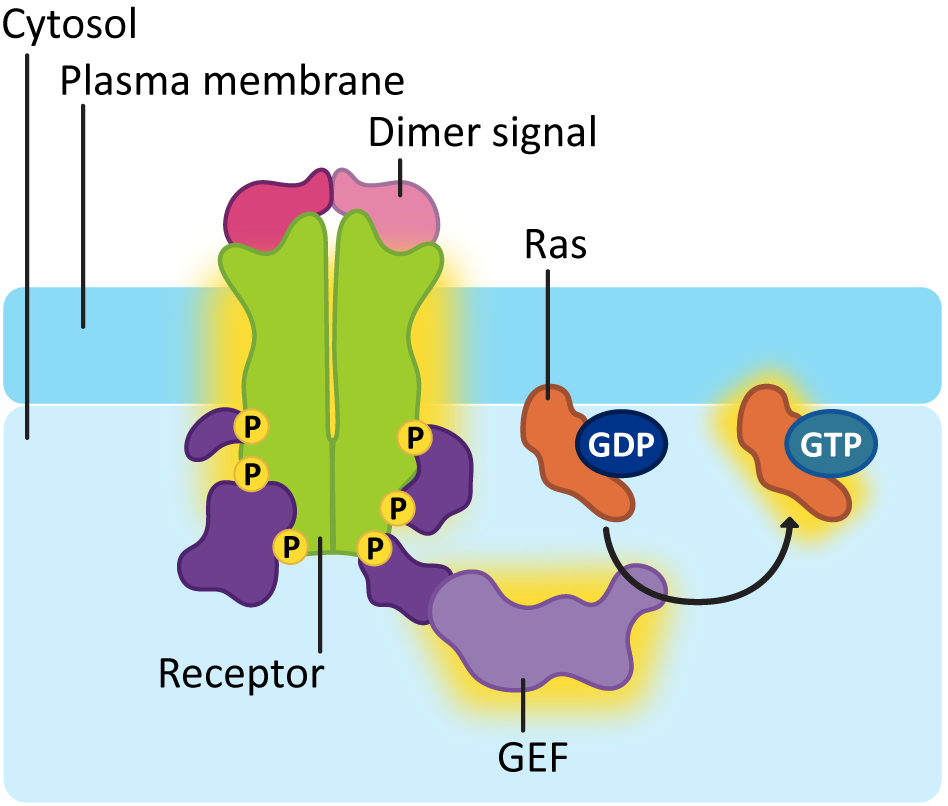
In order for Ras to be activated, a second enzyme known as a guanine-exchange factor, or GEF, is used to swap out the GDP for GTP. As you can see in Figure 07-13, the activation of the receptor allows the GEF to dock, which, in turn, makes sure it’s in the right spot to activate Ras.
Ras activation is also short lived, like G-proteins. The GTP that is bound to the activated Ras will eventually be hydrolyzed with the help of another protein known as GTPase-activating protein, or GAP. One of the more common downstream effects of Ras is that it activates a cascade of kinases that phosphorylate each other. You can think of it as very similar to a domino effect, where one domino knocks down the next, which knocks down the next, and so on. In this case, however, serine/threonine kinases are being activated in sequence, as each one phosphorylates the next. We call this a MAP kinase cascade (Figure 07-14).
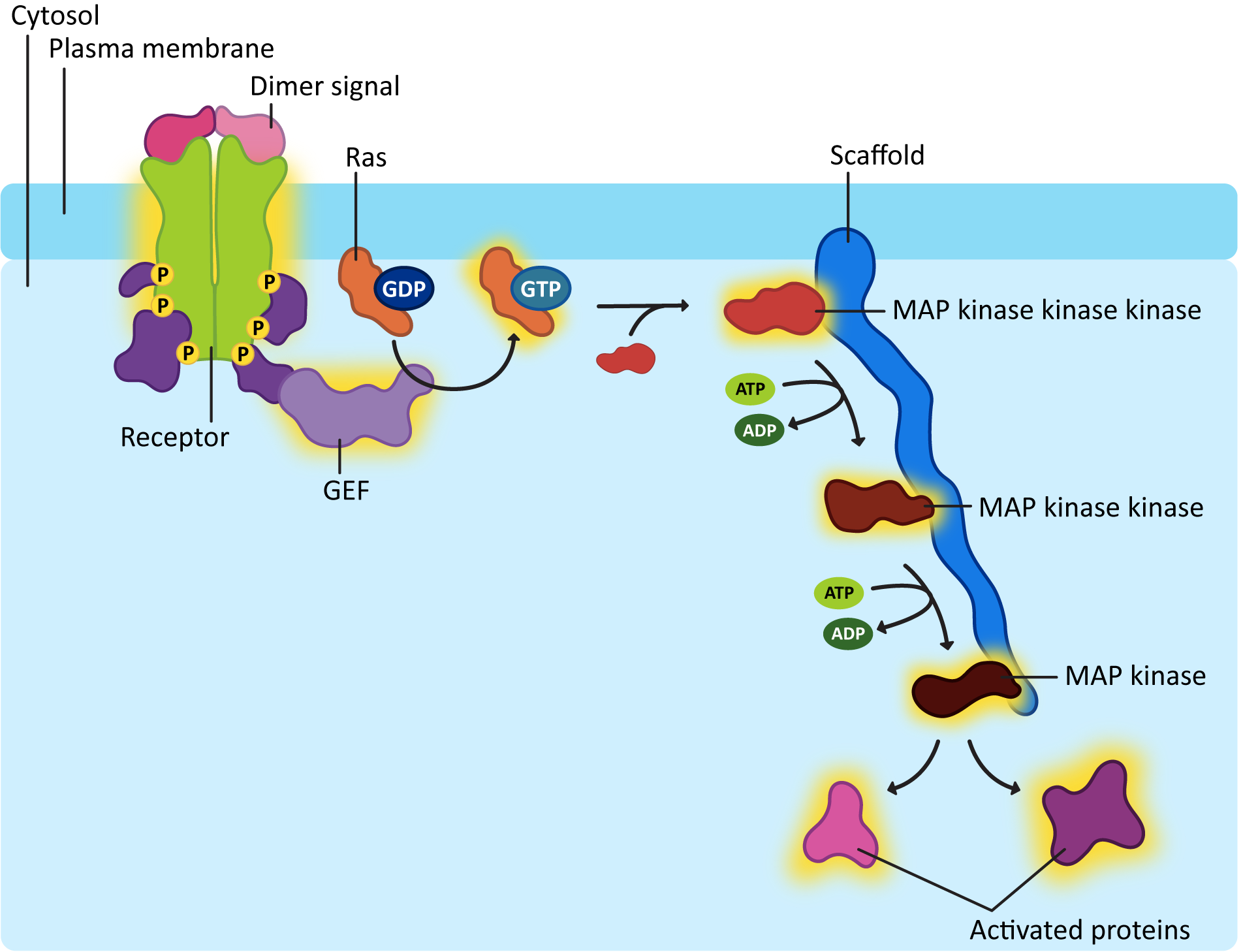
To be quite frank, MAP kinase cascades are very confusing. There are a lot of proteins involved, and the names are not really very creative, which makes things worse. Each of the kinases that gets activated in the cascade has the capacity to go on and activate many other proteins and enzymes, effectively amplifying the signal inside the cell. Usually, scaffold proteins help hold together the correct MAPK proteins. With so many of them in the cell and the fact that they are structurally similar, it would be easy to activate the wrong ones without taking specific measures to avoid that.
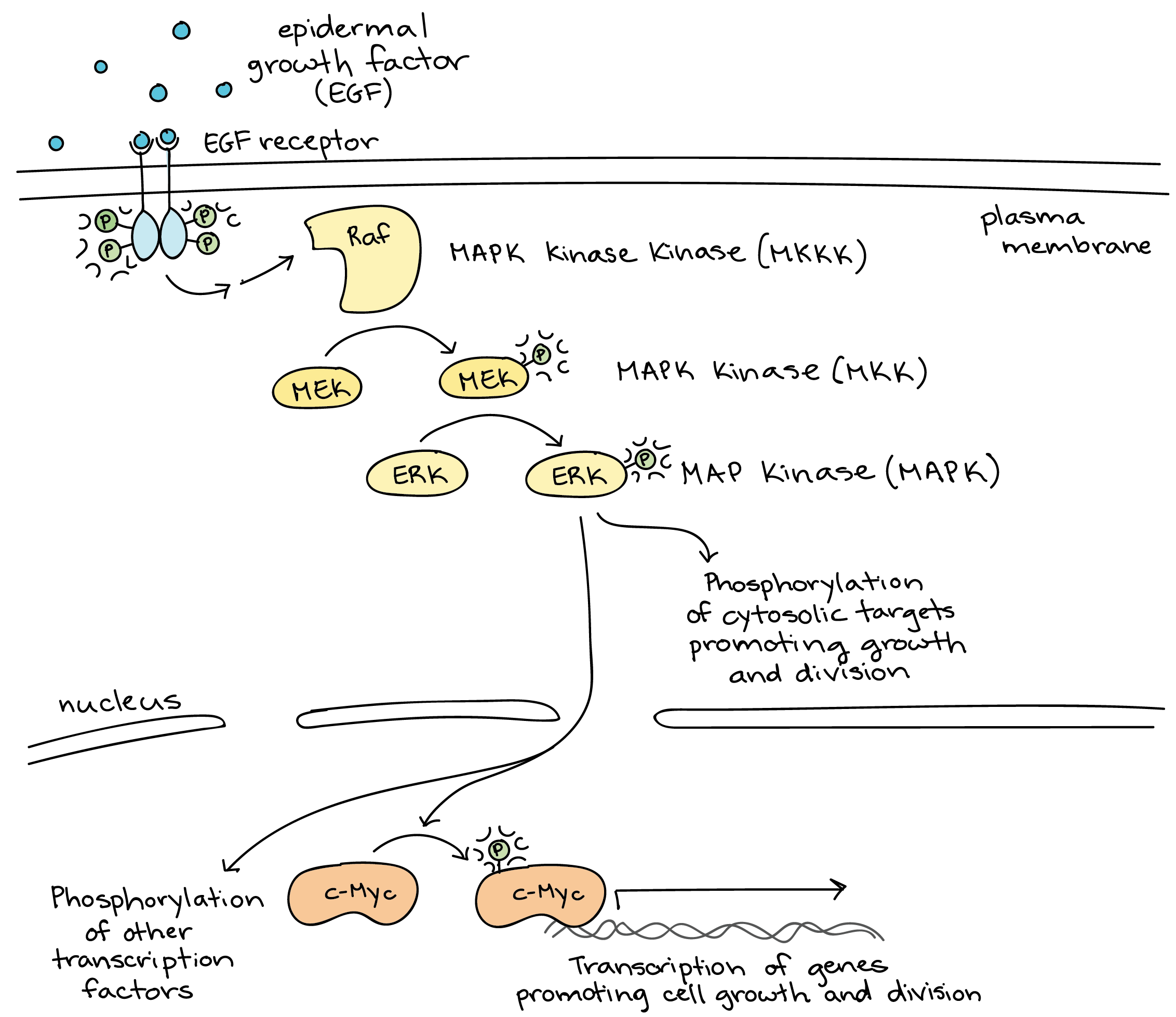
The downstream targets that get activated by a MAP kinase cascade will be involved in many different aspects of cellular function and are likely to have both fast and slow responses, much like all of the rest of the signaling we’ve seen so far. To show you how these cascades can be used, here is an example of a MAP kinase cascade simulated by the growth factor EGF (Figure 07-15). We are lucky to also have a video representation of this cascade to help us visualize what’s going on (Video 07-06).
Downstream Effects of Enzyme-Coupled Receptors—PI 3-Kinase
Another common mechanism that gets activated in an RTK pathway is an enzyme called PI-3-kinase. Phosphatidylinositol (PI) is a type of phospholipid that we’ve seen before. These phospholipids have a 6-carbon sugar (called inositol) as the head group. As we learned earlier in this topic, IP3 and DAG are also produced from a phosphatidylinositol known as PIP2. The reason these particular phospholipids are so useful in signaling is that the inositol ring is exposed at the surface of the membrane and has three different sites exposed where phosphates can be added to it. As a result, you can have PIs with one, two, or three phosphates added in many different combinations. In addition, it is easy to switch the locations of the phosphate groups around. The job of PI-3-kinase is to add a phosphate group at the third carbon in the inositol ring (Figure 07-16).
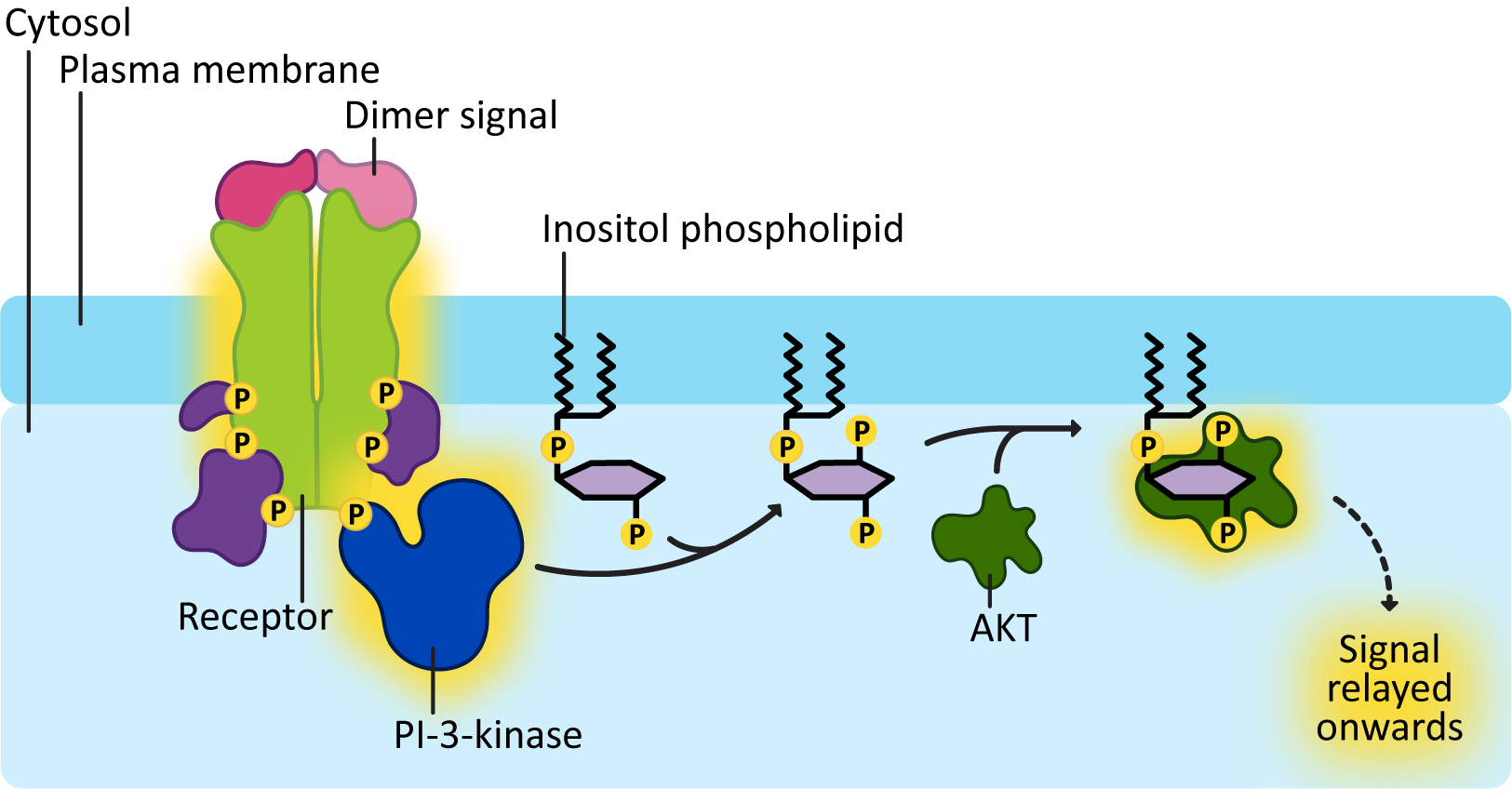
The addition of the phosphate at the third carbon results in the production of specific docking sites for a variety of proteins. For example, Figure 07-16 shows a protein called AKT docking on the modified PI molecule. AKT is a signaling protein specifically involved in cell survival and suppression of programmed cell death. PI-3-kinase gets involved in all kinds of cellular functions, from the formation of vesicle budding sites, to growth and proliferation, to cell motility.
There Is Extensive Crosstalk between Pathways
The final point that is important to discuss, before we move on to experimental techniques, is a reiteration of how complex signaling really is. These pathways and mechanisms that we discuss do not function in isolation. Each and every cell on the planet is in a constant state of sending, receiving, and responding to signals from its environment. In fact, a cell that stops receiving signals from the environment for whatever reason will immediately die, as there will be nothing to stop programmed cell death (a.k.a. apoptosis) from occurring.
Very few signaling pathways are linear. They are more like an intricate and interconnected web, where each part can be influenced by many other parts. One signaling pathway could make another pathway easier or harder to turn on/off. To end this section, we offer an extremely simplified representation of a tiny subset of the signaling that can occur in a cell (Figure 07-17). We encourage you to explore and try to recognize the different parts of the signaling that we have explored in this chapter. Also look for the visual cues of activation and inhibition that we discussed at the end of Topic 7.1, which is indicated in the figure by either a pointed or blunt arrow.

Once you’ve done that, if you’d like to see a more realistic version of the complexity of signaling, the Cell Signaling Technologies website has an excellent set of signaling pathways that you can explore. (Hint: We recommend starting with insulin signaling or the regulation of actin dynamics, as they have most/all of the components we’ve explored, but there are lots of options.) It’s excellent practice to look at pathways you’ve never seen before and try to figure out what you can recognize. Once you start being able to recognize the various parts and proteins of a signaling pathway, it becomes a little bit less scary.
Studying Cells: Investigating the Effects of Phosphorylation in Signaling
You may remember from earlier in the chapter that phosphorylation and dephosphorylation of proteins are very common posttranslational modifications that are used in signaling to regulate protein activity. You may also remember that phosphate groups are added to amino acids by kinases using ATP as the source of both the activation energy and the phosphate group. Most commonly, kinases will add phosphate groups on one of three different amino acid side chains: serines, threonines, and tyrosines (Figure 07-18). In signaling, phosphorylation of the amino acid residues in preexisting proteins allows for quick responses to external cues, which can be easily reversed by later removing the phosphate.
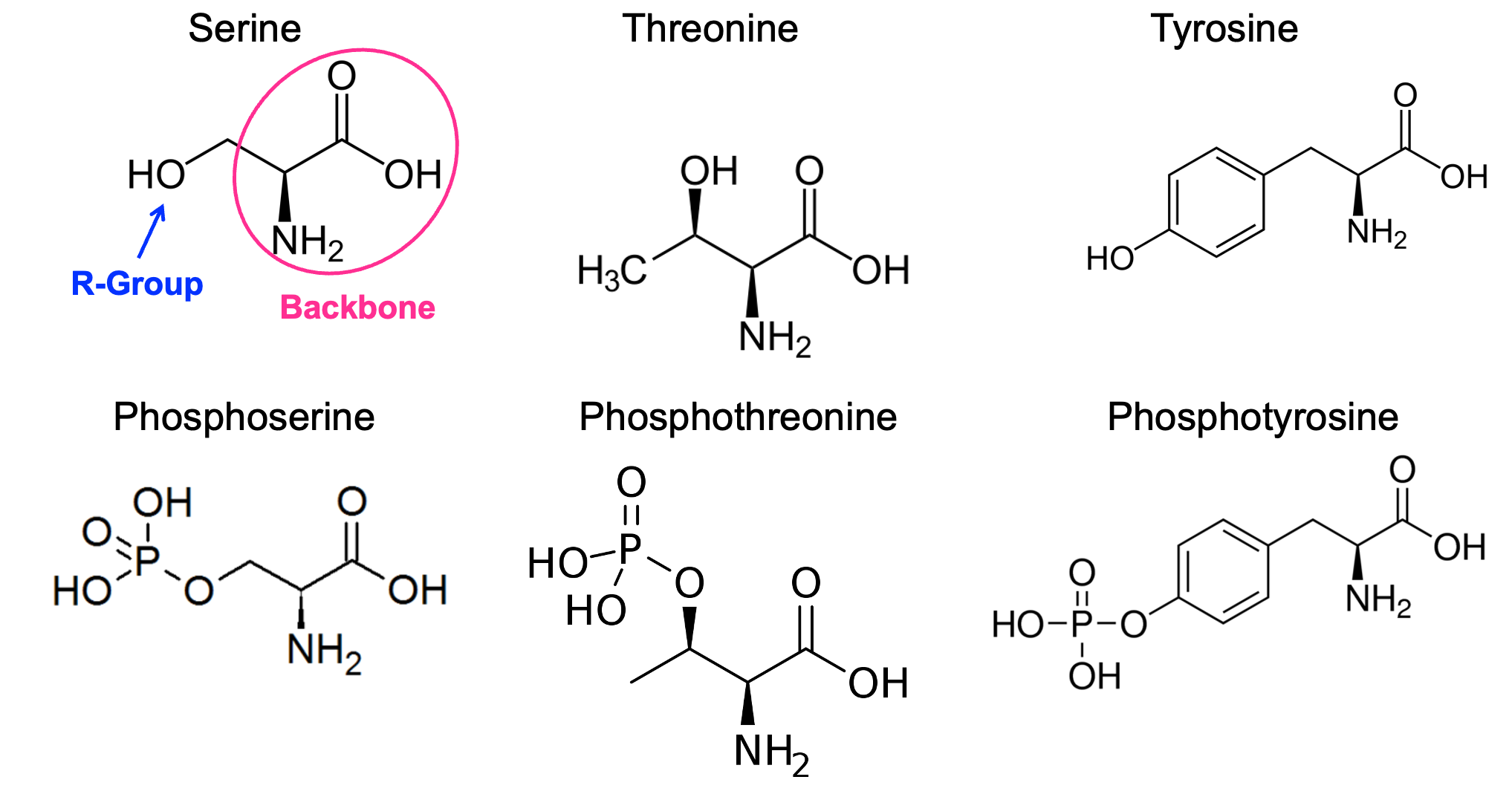
Signaling researchers spend quite a lot of time trying to understand the function of each phosphorylation site on a protein. Dissecting the contribution of an added phosphate to a protein’s overall activity is technically challenging. Bioinformatic analysis has given us the ability to know how many serines, threonines, and tyrosines are present in the protein sequence. However, it cannot tell us which of them are modified by kinases and what the effect of that modification would be on the function of the protein. That’s where experimentation comes in. We will explore two different approaches that are currently use by scientists: phosphomimetics and genetic code expansion (GCE).
Phosphomimetics Attempt to Copy Specific Conformations of Phosphorylated Proteins
In this approach, cell and molecular biologists genetically engineer proteins that have been modified in a particular way using a technique called site-directed mutagenesis. They can use site-directed mutagenesis to specifically replace an amino acid that would normally get phosphorylated with a different amino acid intended to “mimic” the phosphorylated state. Most commonly, negatively charged amino acids are used to replace the phosphorylated amino acid, such as aspartic acid (Asp) or glutamic acid (Glu). The negatively charged Asp and Glu residues are somewhat similar in size and charge to the phosphorylated amino acid, as can be seen in the example in Figure 07-19. The altered proteins have the added advantage that they cannot be dephosphorylated like a phosphorylated amino acid could be. As such, they remain in the “phosphorylated” conformation, which gives us a chance to study the effects of the “phosphorylated” protein in the cell.
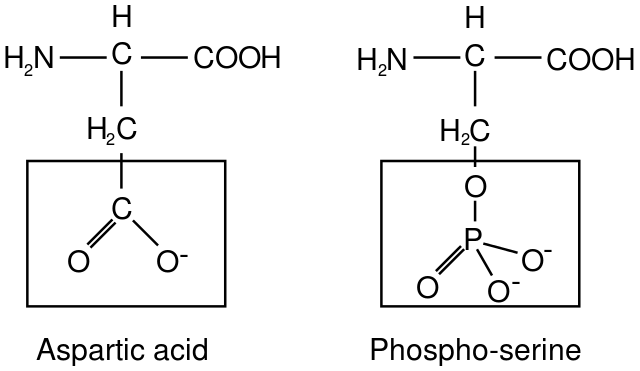
We can also mimic the dephosphorylated form of the protein by genetically engineering an alanine substitution at the site where a serine or threonine would have been originally. Since alanine cannot be phosphorylated, it blocks phosphorylation at that site and “mimics” the unphosphorylated version of the protein.
While these approaches have proven valuable in probing the biological function of phosphorylation of specific residues in some proteins, these approaches also have limitations. As you know from Chapter 2, the primary amino acid sequence determines folding and structure. Replacing one amino acid with another will never be a perfect replication of either the phosphorylated or unphosphorylated form of the original amino acid. The chemical properties of aspartic and glutamic acid are significantly different from serines, threonines, and tyrosines in a variety of ways, including electronegativity, volume, polarity, and hydrogen bonding. All of these chemical properties contribute to how a protein will fold and thus how it will function. Further, the nonphosphorylatable alanine amino acid has a much smaller R group than any of the amino acids it’s meant to replace, and since it’s nonpolar, it lacks their capacity to form hydrogen bonds. Thus, a different approach was devised.
Genetic Code Expansion (GCE) Allows Us to Code for Phosphorylated Versions of Amino Acids
To get around the problems that can arise with phosphomimetics, researchers created a new way to build phosphorylated versions of proteins without requiring the use of amino acids that “mimic” the original function. GCE is a technique that allows us to treat the phosphorylated version of the amino acid as its own entity within the genetic code so that we can insert it directly into the protein sequence. GCE is still a relatively new technique, but it is proving to be very powerful as we seek to learn more about posttranslational modifications, such as phosphorylation, and their role in protein function. Oregon State University is a world leader in this technique through their research center, the GCE4All Center.
The essence of this technique depends on how cells read and interpret the genetic code. The idea is that we “hijack” and modify how the cells interpret specific codons of the genetic code so that they now code for new, unique amino acids. Of course, the genetic code has 64 codons that code for 20 amino acids, and it is interpreted virtually identically across the entire tree of life. However, there are exceptions, most commonly in bacteria, where some codons are interpreted differently and code for different, unique amino acids. In these organisms, we have a different, expanded set of amino acids with which to create proteins. In GCE, we genetically engineer this exact same scenario to include specific new amino acids of our own choosing. For example, we could hijack a codon to code for phosphoserine instead of regular serine so that a phosphorylated version of the protein can be translated directly.
While the premise for GCE is relatively straightforward, hijacking the codons and making them code for a different amino acid requires careful manipulation of several components of the protein translation machinery. Remember that a key component of proper protein translation is the correct addition of the amino acid to the tRNA (for a refresher on translation, see this link from the Khan Academy). So if we want to change which amino acid a particular codon codes for, we must find a way to change which amino acid is attached to that codon. Adding the amino acid to a specific tRNA is done by a family of proteins known as amino acyl tRNA synthetases. Changing the genetic code begins with genetically modifying these synthetases so that the amino acid that is added to the tRNA is altered. Figure 07-20 provides a visual representation of this. In the figure, one of the stop codons (UAG) has been repurposed to code for phosphoserine (pSer in the figure). Once you have created this machinery, you can use site-directed mutagenesis to add your newly repurposed codon in any site in the genetic sequence where you want the new amino acid to be added, and the translation machinery will do the job for you. It’s worth noting that this system is mostly done in test tubes (i.e., in vitro), as changing the codon language used to create every protein in a cell would likely be detrimental and kill the cell.
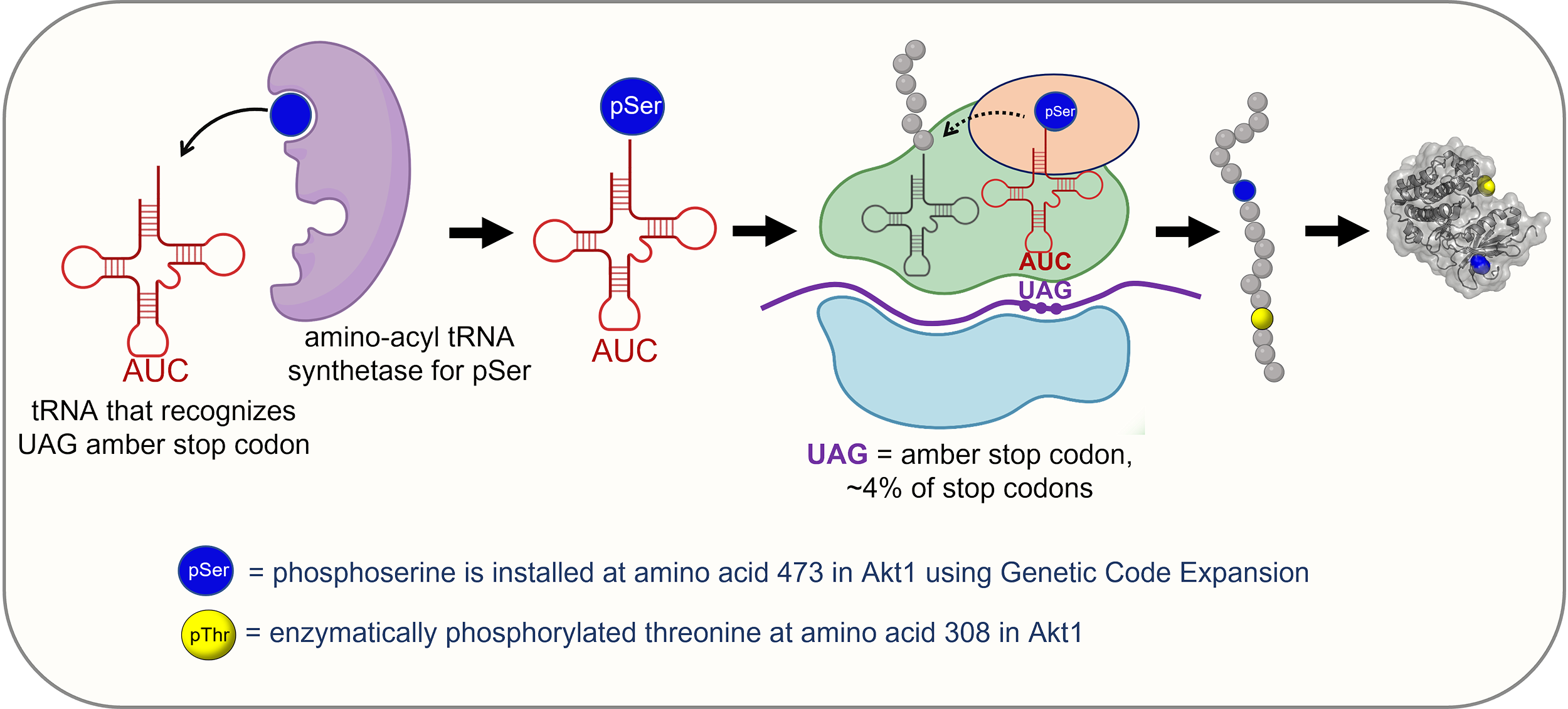
The advantage of this technique, compared to using phosphomimetics, is that you can add an actual phosphoserine rather than using a closely approximated amino acid, such as aspartic acid. Researchers do not know which kinases are involved in phosphorylation of a protein at a particular site. Using this method, they can create a phosphorylated version for use in vitro and then examine the protein’s activity by looking at its effect on known downstream targets.
GCE can be and has been used to address a wide variety of research questions that go far beyond the phosphorylation research described here. Many research areas have benefited from GCE, such as cell signaling, disease regulation, protein localization, protein-protein interactions, therapeutic development, and novel biomaterials and sensors. Its strength is in the fact that the novel amino acids that can be introduced into the genetic code are almost endless as long as the translation machinery can still work with them. They have been used to study topics such as posttranslation modification of proteins, linking two proteins together, adding on a stable fluorescent dye for microscopy in live cells, and other forms of labeling proteins to learn more about their structure. There are some examples of research that have been supported by Oregon State’s GCE4All Center, which gives you an idea of the versatility of this technique.
Chapter Summary
In this chapter, we explored how cells communicate and respond to their environment through cell signaling. In the first topic, we explained the fundamental anatomy of a signaling cascade, from the properties of a signal molecule, to the receptors to which it binds, to the intracellular signaling molecules, and finally to the types of responses that can be elicited from any given signal. Then we delved into specific examples of signaling systems and common pathways you might encounter in past and future coursework. Finally, we explored how we can use a couple of different genetic techniques to study how posttranslational modifications affect signaling cascades to understand the regulation of these complex systems. Hopefully, after reading this chapter, you will agree that this fundamental cellular process is as beautiful as it is complex.
Review Questions
Note on usage of these questions: Some of these questions are designed to help you tease out important information within the text. Others are there to help you go beyond the text and begin to practice important skills that are required to be a successful cell biologist. We recommend using them as part of your study routine. We have found them to be especially useful as talking points to work through in group study sessions.
Topic 7.1: General Principles of Signaling
- We described five major types of signaling used by multicellular organisms. What are they? What are the benefits of each one? How do they impact the affinity of the receptors that are used in this form of signaling?
- Explain the difference in the chemical properties of signals that use internal receptors compared to those that are restricted to using cell-surface receptors.
- Describe the types of responses that can be elicited in cells when a signal is received.
- Compare the pros and cons of fast versus slow response rates for a signaling pathway.
- List the pros and cons of long-range (endocrine) signaling as opposed to short-range (contact or paracrine) signaling.
- Explain/define the following terms and know the function and significance of each:
- cell-surface receptors
- signal transduction
- second messenger
- cell response
- endocrine signaling
- paracrine signaling
- contact-dependent signaling
- synaptic signaling
Topic 7.2: Examples of Common Signaling Mechanisms
- Describe the activation of an ion-gated channel.
- Explain why it can be said that ion-gated channels convert chemical energy into electrical energy.
- Describe activation of a G-protein-coupled receptor (GPCR) pathway from the binding of a signal at a GPCR to the activation of downstream signaling molecules.
- Name and describe common potential downstream signaling molecules/events in a GPCR pathway.
- Explain/define the following terms, know which ones are the same, and know the function of each:
- GPCR
- heterotrimeric G-protein
- hydrolyzation (hydrolysis)
- IP3
- DAG
- adenylyl cyclase
- CAMP (cyclic AMP)
- phosphomimetic
- phosphorylation
- posttranslational modification
- genetic code expansion (GCE)
- receptor tyrosine kinase
- autophosphorylation
- Ras, structure and features of Ras
- MAP kinases
- scaffold proteins
- Name some examples of signal molecules that bind to receptor tyrosine kinases.
- Explain the events following the binding of a signal to a receptor tyrosine kinase and how these events bring about change in cells.
- Name and describe common potential downstream signaling molecules/events in a receptor tyrosine kinase pathway.
- Describe a mechanism that allows the specificity of signaling pathways despite redundancy of signaling molecules.
- Describe how phosphorylation is an important posttranslational modification in signaling pathways.
- Explain how phosphorylation affects proteins chemically.
- List some pros and cons of using phosphomimetics or GCE to study the role of phosphorylated proteins in signaling pathways.
A long-range signaling pathway where a cell secretes a signaling molecule that is picked up by target cells not in the local vicinity.
Referring to neurons, an electrically excitable cell that is associated with the brain.
A short-range signaling pathway where a cell secretes a signaling molecule that is picked up by target cells in the local vicinity.
In this type of signaling, the signal is released and received by the same cell.
In this contact dependent signaling pathway, a cell attaches to another cell to initiate the signaling pathway.
A protein that binds a signal molecule to transmit a cascade intracellularly.
In the context of proteins, refers to the favorability of a binding interaction. For instance, the protein when phosphorylated has greater affinity for its substrate. This means that the protein is likely to bind more tightly when the protein is phosphorylated.
A molecule that binds to a receptor to initiate a signaling cascade.
“A class of signaling molecules in multicellular organisms that are sent to distant organs by complex biological processes to regulate physiology and behavior.” Source: Hormone. (2023, July 24). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Hormone
“A signaling molecule secreted by a neuron to affect another cell across a synapse.” Source: Neurotransmitter. (2023, April 17). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Neurotransmitter
A regulated process of cell death.
The chemicals that make up smell molecules. These are detected by odorant receptors in the nose. Subsequent signaling and activation of neurons to the brain help us sense these molecules.
“In mammals, including humans…a signaling molecule in many physiological and pathological processes.” Source: Nitric oxide. (2023, July 11). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Nitric_oxide
“A class of proteins responsible for sensing steroids, thyroid hormones, vitamins, and certain other molecules. These intracellular receptors work with other proteins to regulate the expression of specific genes thereby controlling the development, homeostasis, and metabolism of the organism.” Source: Nuclear receptor. (2023, May 25). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Nuclear_receptor
Refers to the process of signal transduction. When a signal is transduced, it is changed from its original form along the pathways. For example, when a signal binds to a receptor, this induces a conformational change in the receptor to activate it. The signal can be said to be transduced from a physical molecule to an activated receptor.
“A series of chemical reactions that occur within a biological cell when initiated by a stimulus.” Source: Biochemical cascade. (2023, June 16). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Biochemical_cascade
“The process by which a chemical or physical signal is transmitted through a cell as a series of molecular events.” Source: Signal transduction. (2023, June 20). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Signal_transduction
The attachment of a phosphate group to a biological molecule. When applied to a protein, this can often change the conformation of the protein, resulting in a functional change.
The process of removing a phosphate chemical group from a protein.
Proteins that “interact and/or bind with multiple members of a signaling pathway, tethering them into complexes. In such pathways, they regulate signal transduction and help localize pathway components (organized in complexes) to specific areas of the cell.” Source: Scaffold protein. (2021, September 17). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Scaffold_protein
Biological molecules released as part of a signaling cascade as a way to amplify the signal.
A regulatory mechanism whereby the product of a process will influence (upregulate or downregulate) itself.
A regulatory process where a receptor is brought into the cell via endocytosis, and then degraded in the lysosome.
A regulatory process where a receptor is endocytosed such that it isn’t available to bind to signal molecules on the cell’s surface.
A regulatory process where a biological molecule directly inhibits the cell receptor’s function.
When a biological molecule that is part of a signaling cascade is prevented from participating in its role.
The region 5′ of the indicated location in a piece of DNA.
Refers to the region of the DNA that is 3′ of the area referenced. For example, in a sentence, you could say, “The transcription stop site is located downstream of the transcription start site.”
When a signal binds to this type of receptor, an ion channel is opened as a result.
“An inositol phosphate signaling molecule. It is made by hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2), a phospholipid that is located in the plasma membrane, by phospholipase C (PLC). Together with diacylglycerol (DAG), IP3 is a second messenger molecule used in signal transduction in biological cells.” Source: Inositol trisphosphate. (2023, May 1). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Inositol_trisphosphate
“Phosphatidylinositol 4,5-bisphosphate or PtdIns(4,5)P2, also known simply as PIP2 or PI(4,5)P2, is a minor phospholipid component of cell membranes. PtdIns(4,5)P2 is enriched at the plasma membrane where it is a substrate for a number of important signaling proteins.” Source: Phosphatidylinositol 4,5-bisphosphate. (2023, January 26). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Phosphatidylinositol_4,5-bisphosphate
“A large group of evolutionarily related proteins that are cell surface receptors that detect molecules outside the cell and activate cellular responses. They are coupled with G-proteins.” Source: G-protein-coupled receptor. (2023, April 16). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/G_protein-coupled_receptor
“A family of proteins that act as molecular switches inside cells and are involved in transmitting signals from a variety of stimuli outside a cell to its interior. Their activity is regulated by factors that control their ability to bind to and hydrolyze guanosine triphosphate (GTP) to guanosine diphosphate (GDP).” Source: G-protein. (2023, July 21). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/G_protein
“A second messenger, or cellular signal occurring within cells, that is important in many biological processes. cAMP is a derivative of adenosine triphosphate (ATP) and used for intracellular signal transduction in many different organisms, conveying the cAMP-dependent pathway.” Source: Cyclic adenosine monophosphate. (2023, April 30). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Cyclic_adenosine_monophosphate
“A glyceride consisting of two fatty acid chains covalently bonded to a glycerol molecule through ester linkages.” Source: Diglyceride. (2022, November 2). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Diglyceride
An enzyme responsible for making cAMP (cyclic AMP) from ATP. Also called adenylate cyclase.
An enzyme that cleaves a phosphodiester bond. These occur in between nucleic acids or within a ringed nucleotide such as cyclic AMP.
“A family of serine-threonine kinase whose activity is dependent on cellular levels of cyclic AMP (cAMP).” Source: Protein kinase A. (2023, July 23). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Protein_kinase_A
“Phospholipase C’s role in signal transduction is its cleavage of phosphatidylinositol 4,5-bisphosphate (PIP2) into diacyl glycerol (DAG) and inositol 1,4,5-trisphosphate (IP3), which serve as second messengers.” Source: Phospholipase C. (2023, June 24). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Phospholipase_C
A regulatory mechanism whereby the product of a process will negatively influence (downregulate) itself.
Kinases (enzymes that add a phosphate group) on serine or threonine amino acids of the target protein.
An enzyme that adds a phosphate chemical group to another enzyme. These can be added on serines, threonines, or tyrosine.
Proteins that remove a phosphate from a target protein.
“The high-affinity cell surface receptors for many polypeptide growth factors, cytokines, and hormones.” Source: Receptor tyrosine kinase. (2022, February 19). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Receptor_tyrosine_kinase
The process of two biomolecules binding.
Protein locations where other biomolecules are able to bind.
“A family of related proteins that are expressed in all animal cell lineages and organs. All Ras protein family members belong to a class of protein called small GTPase, and are involved in transmitting signals within cells.” Source: RasGTPase. (2023, March 17). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Ras_GTPase
“Proteins can switch between active and inactive states, thus acting as molecular switches in response to another signal. For example, phosphorylation of proteins can be used to activate or inactivate proteins.” Source: Molecular switch. (2023, June 10). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Molecular_switch
Existing as a single unit. With a protein, this refers to having a single polypeptide chain.
A family of proteins that will swap a GDP for GTP on a G protein.
A family of proteins that helps a G protein accelerate its GTPase activity (converting GTP into GDP).
A three-part signaling cascade where a MAPKKK phosphorylates a MAPKK, which then phosphorylates a MAPK. These are part of a larger signaling pathway.
“A family of related intracellular signal transducer enzymes capable of phosphorylating the 3 position hydroxyl group of the inositol ring of phosphatidylinositol (PtdIns).” Source: Phosphoinositide 3-kinase. (2022, October 27). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Phosphoinositide_3-kinase
A ringed sugar molecule that is commonly used in signaling pathways.
An amino acid substitution in a engineered protein to mimic the rough charge and shape of a phosphorylated protein at that location.
A biochemical technique to encode designer amino acids into genetically engineered proteins.
Responsible for putting the amino acid on the tRNA.

