2 Biological Membranes
Introduction
Now that you’ve had a chance to better understand microscopes, we now turn to learning about the makeup of the cells themselves. In this chapter, we will start by exploring the composition and function of biological membranes. This will give us the opportunity to review what we know about macromolecules, specifically lipids and proteins and, to a lesser extent, carbohydrates. After that, we will discuss the function of the plasma membrane and how the structure of those macromolecules contributes to the function of the plasma membrane. Finally, we will discuss cell surface–adjacent structures, like the extracellular matrix or plant cell wall, that are found just outside of the plasma membrane.
Topic 2.1: The Chemical Features of Biological Membranes
Learning Goals
- List the four primary features of a biological membrane and explain why they are important for cellular function.
- Explain how the chemical composition of a membrane (including lipids, carbohydrates, and proteins) contributes to its function.
- Explain how thermodynamics and the hydrophobic effect hold membranes together.
- Compare and contrast the chemical properties of membranes with the molecules that are allowed to cross versus the ones that are excluded from crossing.
All membranes in the cell share some common features, despite the fact that they each have a different composition, fluidity, and permeability. In this topic, we will discuss the common features of all membranes and touch briefly on which macromolecules contribute to specific membrane functions.
It’s worth noting that a biological membrane is not the same thing as a phospholipid bilayer, despite the fact that these terms sometimes are used interchangeably. A phospholipid bilayer is made only of phospholipids and nothing else, whereas a biological membrane will have many types of lipids in it (including glycolipids and cholesterol) as well as proteins. We’ll see examples of this later in the topic.
Membranes Are Barriers That Define Compartments
One of the most basic features of cells is that they are separated from the environment by a membrane. Not only do the membranes create a barrier that separates the inside of the cell from the exterior, but they also function in a way that gives the cell a great deal of control over what can enter or exit it. This creates an environment inside the cell that is different from the outside (Figure 02-01).
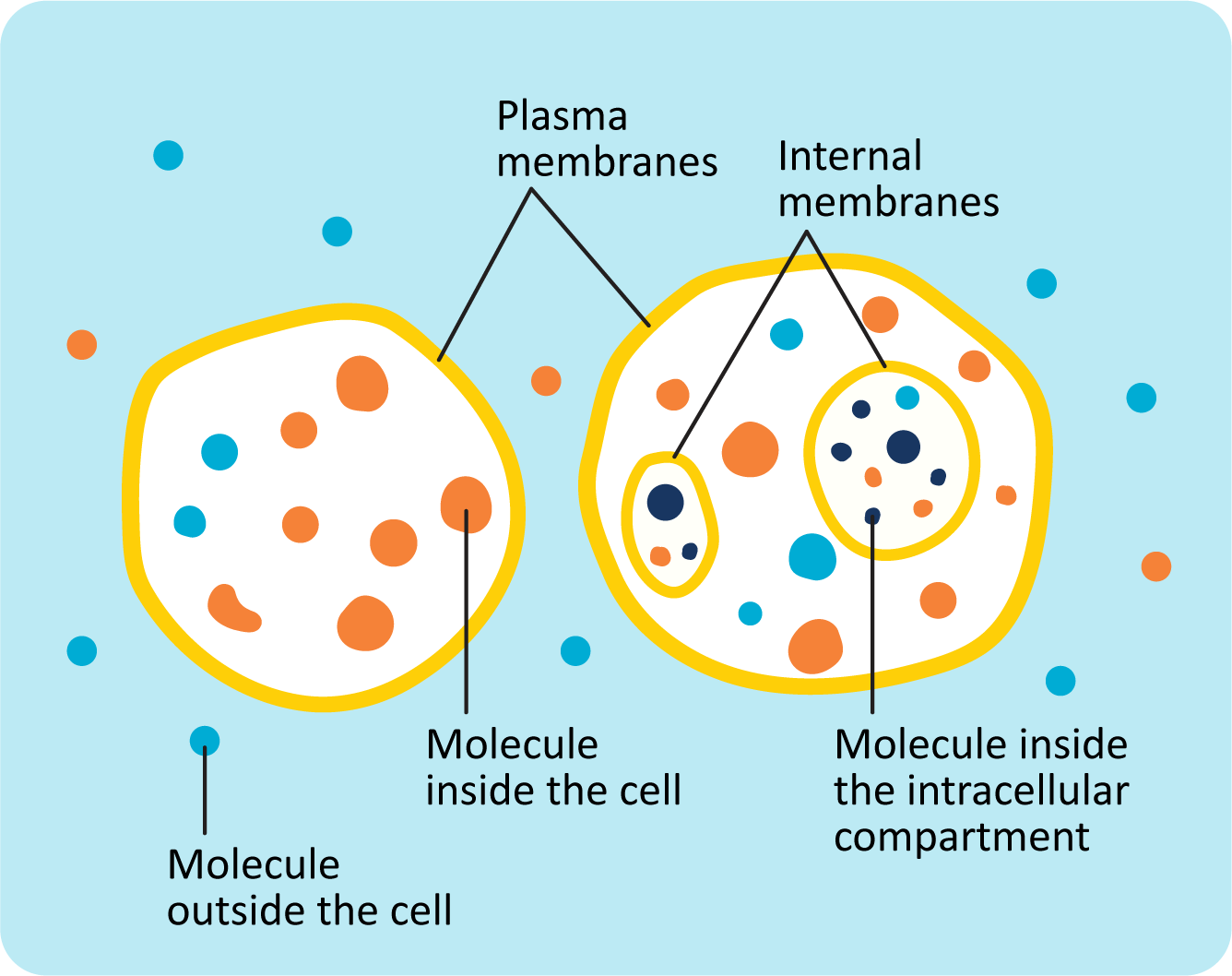
Molecules are able to move around freely (diffuse) within each cellular compartment. However, it is more challenging for some molecules to cross membranes. This is because biological membranes have specific chemical properties, which determine when and how molecules cross the membrane. Thus, membranes are a key feature that allows cells to maintain compartments with a distinctive chemical composition.
Like everything in science, the idea that biological membranes contain both lipids and proteins took time for cell biologists to accept. Sometime in the 1970s, the current concept of the membrane was developed and was known as the “fluid mosaic model.” The “fluid” part refers to the lipids being able to move around within the layers of the membrane, and the “mosaic” part refers to the fact that proteins are expected to be scattered across and throughout the membrane. Since then, further research and experimentation have expanded and refined our understanding of how membranes work. In this part of the chapter, we will explore four general features of biological membranes:
- The membrane is a bilayer, made up of lipids and proteins.
- The membrane is selectively permeable.
- The membrane is organized but fluid.
- The membrane is asymmetric.
We will look at each of these in a little more detail. Then in Topics 2.2 and 2.3, we will take a more in-depth look at the relationship between lipids and membrane fluidity as well as the organization and function of membrane proteins.
General Features of Biological Membranes
The Membrane Is a Bilayer Made Up of Both Lipids and Proteins
The earliest work on membranes was done on plasma membranes from red blood cells. This is because red blood cells are easy to collect in relatively large quantities. In addition, in mammals, they lack a nucleus and internal membranes. So they contain mostly “pure” plasma membrane without the need to experimentally separate other membrane types during experimental isolation. To isolate these membranes, cells are burst open and the membranes are gently washed to remove cytoplasmic debris. Lipids and proteins are separated by treatment with strong detergents or organic solvents (e.g., ether) and characterized. Next, we’ll take a look at the chemical properties of lipids and proteins separately to discuss how they each contribute to the structure of the biological membrane.
1a. The Lipid Component of Membranes: Formation of Lipid Bilayers
(For a complete review of the basic structure and formation of lipid molecules as well as a description of polar and nonpolar molecules, please see the introduction.)
The main lipid components of membranes are phospholipids, a family of molecules with a polar phosphate head group and two fatty acid tails, each connected to an arm of a glycerol via an alcohol residue (Figure 02-02A). However, they are almost never alone in a biological membrane. Other lipids (glycolipids, sphingolipids, and cholesterol) as well as a great many proteins are found in every biological membrane. The chemical properties of phospholipids allow them to adopt a bilayer shape, a key component to a biological membrane.
In water, fats and oils will form into large droplets of jumbled molecules that do not mix with the water (think salad dressing). However, membrane lipids are able to form sheets because of two very important characteristics:
- They are amphipathic (i.e., there is a polar “head” region, which can freely form hydrogen bonds with water, and a nonpolar “tail” region, which cannot). Some sources refer to polar molecules/regions as hydrophilic, which translates to “water loving.” Conversely, the nonpolar molecules/regions are sometimes referred to as hydrophobic, which translates to “water fearing,” but as these terms are misleading and stray from referencing the chemical properties of the molecules, we don’t find them to be as useful. We prefer the terms polar and nonpolar when describing the head and tail regions of the phospholipids, as they are more scientifically accurate. However, both will be used in this textbook.
- They form a roughly cylindrical shape, which tends to stack together well (much like cans of soda; see Figure 02-02B).
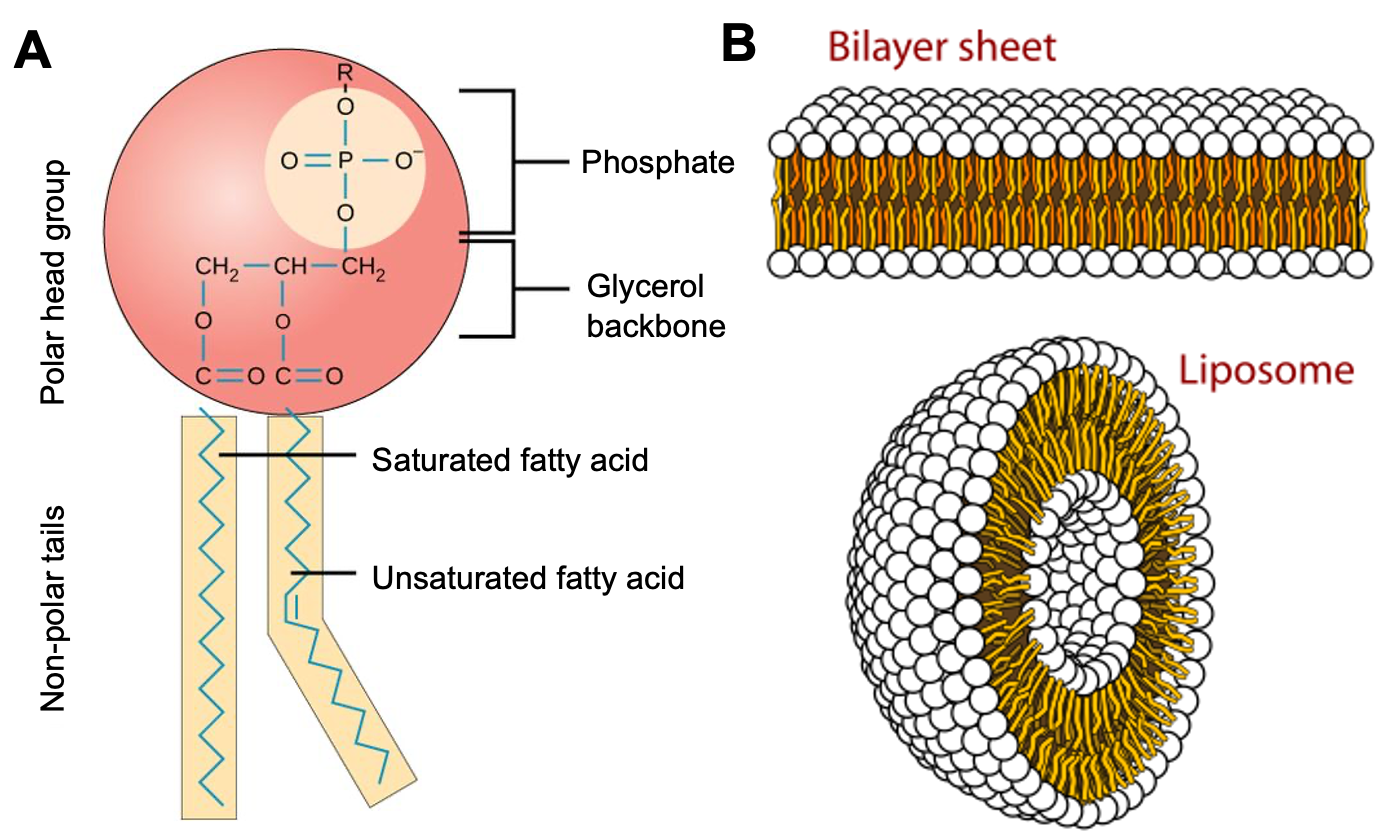
Phospholipids, such as the one shown in Figure 02-02A, come together to form a bilayer—two sheets of phospholipids with their polar head groups oriented outward. Each layer of the bilayer is called a leaflet. Since the nonpolar tails of the phospholipids face inward, away from the water, they are sequestered away and do not interact directly with the water molecules (Figure 02-03). This orientation allows for the maximum freedom of the surrounding water. Because of the stable membrane conformation, if a tear or a hole is created in a lipid bilayer or a biological membrane, it rapidly seals up again.

When phospholipids are mixed with water, they will spontaneously form into spherical bodies called liposomes that have water on the inside as well as on the outside (Figure 02-02B). Liposomes are commonly used for the targeted oral delivery of drugs and other agents in medical treatment. Liposomes are the simplest version of a cell membrane, so they are able to fuse with a real cell membrane to release its contents directly into the cell. This is useful if the compound inside the liposome would not easily pass through a membrane under normal circumstances. A great example of this technology in action is the RNA vaccines developed to target the SARS-CoV-2 virus (the cause of the COVID-19 pandemic). These RNA vaccines use an outer coating called a lipid nanoparticle, which is a combination of phospholipids, cholesterol, and other compounds that are designed to help contain and stabilize the RNA vaccine and ease its entry into the cell.
Thermodynamics Drive the Formation of the Lipid Bilayer: The Hydrophobic Effect
Phospholipids form stable bilayers in an aqueous environment due to thermodynamics. The spontaneous formation of a lipid bilayer from lipids in an aqueous solution, simulated in Video 02-01, shows the power of thermodynamics in action. The laws of thermodynamics explain why it is more energetically favorable for phospholipids to clump together and form a bilayer in water than it is for them to remain dispersed.
The phenomenon where nonpolar molecules in an aqueous environment clump together is called the hydrophobic effect. Clumping together is not favorable because the nonpolar groups attract each other per se, but because, when clumped together, nonpolar molecules do not have to interact as much with water, which is polar. When the nonpolar material clumps together, water and other polar molecules are freer to move and, more importantly, hydrogen bond with each other. This freedom of motion for the water allows for the overall reaction to be spontaneous.
While we don’t want to get too far into the equations of thermodynamics, here is a brief description of how thermodynamics favors the hydrophobic effect. You may want to return to your general chemistry notes for this (or refer back to the information in the introduction).
Recall from general chemistry that the free energy equation is
[latex]\Delta G= \Delta H-T \Delta S\text{,}[/latex]
where ΔG is the Gibb’s free energy change, ΔH is the change in enthalpy (often described as internal energy or bond energy of the molecule), ΔS is the change in entropy (which is often thought of as motional freedom), and T is temperature.
As a reminder, reactions are energetically favorable when ΔG is negative, which means that energy will be released. This can be accomplished by decreasing the enthalpy or by increasing entropy. In the case of the formation of the lipid bilayer (and any other clustering of nonpolar molecules in water), it is the entropy of the water molecules surrounding the nonpolar groups that is changed when lipids assemble into bilayers. Two points are critical here:
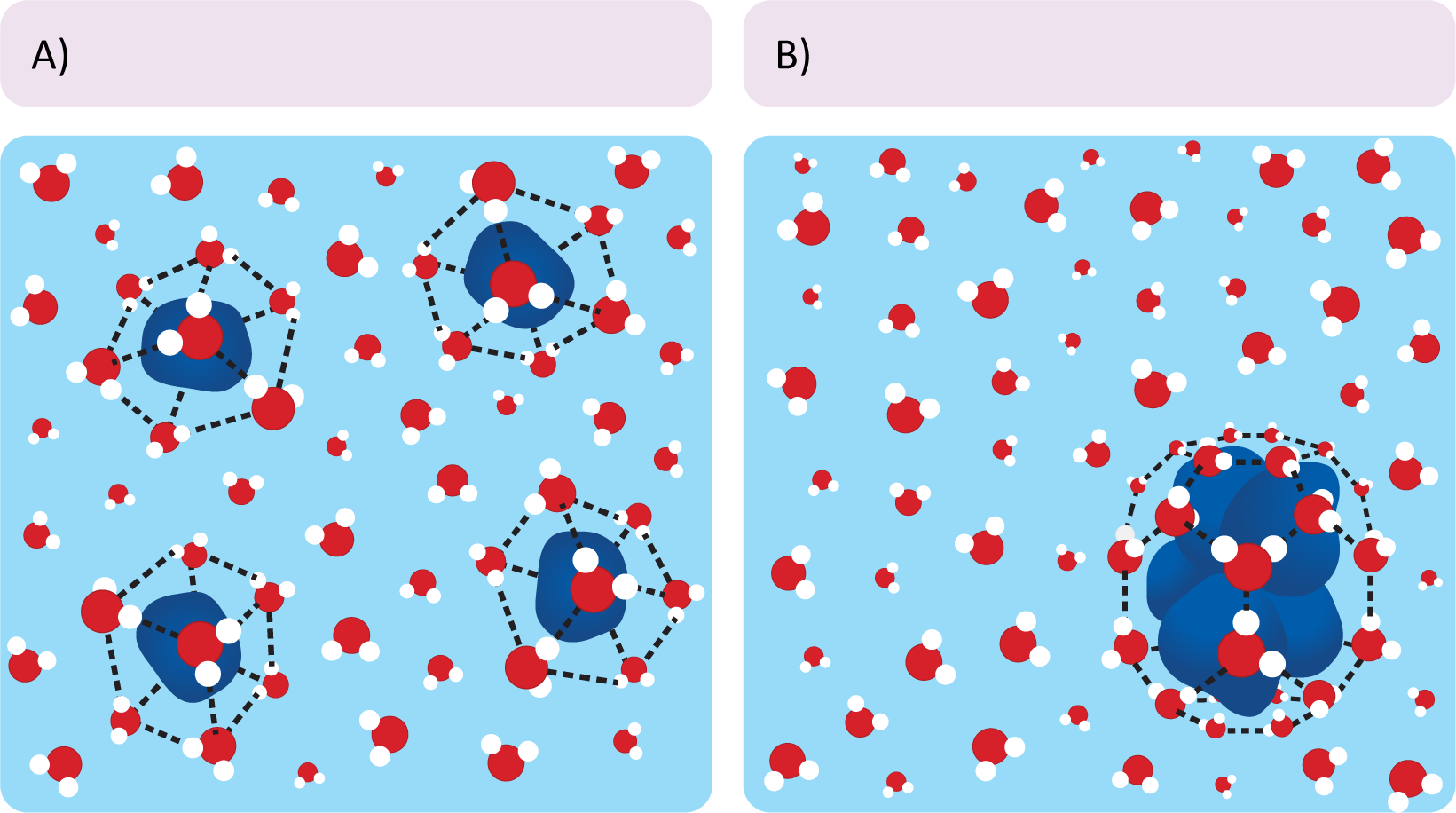
- Nonpolar particles force the surrounding water molecules into an energetically unfavorable configuration that distorts the normal hydrogen-bonded structure of water (Figure 02-04). This cage-like structure restricts the movement of the water surrounding the nonpolar particle. Clumping the nonpolar molecules together reduces the surface area that is required for the nonpolar molecules to interact with the surrounding water.
- In the case of phospholipids, when the nonpolar lipid tails are sequestered away from the surrounding aqueous environment, they don’t come into contact with water at all. Thus, the water molecules are not constrained and are able to freely hydrogen-bond with the polar head groups and/or other polar molecules. This net reduction of the number of “constrained” water molecules provides a large increase in entropy. Based on our equation above, all other things being equal, that increase in entropy can be enough to result in a negative change in free energy. Thus, it requires less overall energy to hold phospholipids in a bilayer formation, making it “energetically favorable.”
1b. The Protein Component of Membranes
The second major component of membranes is proteins, which should come as no surprise. Virtually everything in the cell is either made of proteins or made by proteins, so the importance of understanding how the structure of proteins impacts their function cannot be overstated.
We expect that you have learned about proteins in your general biology classes, so we approach this topic from that perspective. (If you need a refresher, we suggest looking at the material in the introduction.) However, even if you have learned about proteins before, there is much more to learn. Proteins are important enough that you should expect to cover them in several courses, with increasing levels of detail and complexity. In this textbook we will be exploring protein structure and function, and how it specifically relates to membrane structure and function, in Topic 2.3.
General Features of Biological Membranes
The Membrane Is Selectively Permeable
We say that membranes are selectively permeable because experiments show that they allow some molecules to pass through while excluding others. However, it has also been shown that the properties of different membranes vary…a lot! Some are very permeable to ions and water, while others allow almost nothing through.
The chemical composition of the membrane, specifically the lipids that make up the bilayer, is a large factor in the membrane’s capacity to be selectively permeable. Due to the primarily nonpolar environment inside the core of the bilayer, it is extremely unfavorable for water or other polar molecules to spend any time in there; thus, the interior of the membrane is almost entirely a water-free zone. This does not necessarily mean that small polar molecules (like water) never enter the core of the bilayer, but if they do, they exit again quickly. This is the basis for the reduced permeability of membranes for polar molecules.
To further explore selective permeability, we will look at the simplest example—a synthetic bilayer made entirely of phospholipids. In biological membranes, there are a variety of transport proteins, which allow molecules to pass through the membrane. Looking first at a pure phospholipid bilayer can help us understand why some molecules need transporters, whereas others do not.
- Depending on the properties of the molecule in question (size, polarity), different molecules will have more or less difficulty crossing through that nonpolar portion of the membrane. Small nonpolar molecules (like oxygen, carbon dioxide, and nitric oxide) can cross a lipid bilayer easily, without any help.
- Polar molecules that are very small (like water) are also able to cross bilayers, but not very well. As such, the process is slower, and many membranes have transport proteins to facilitate passage when necessary.
- Larger molecules (amino acids, nucleotides, and glucose) will struggle to cross the bilayer as a result of their size. These molecules can also be polar, which can add to the challenge.
- Charged molecules, regardless of size, will be completely unable to pass through the bilayer without help. This is the basis for many of the electrochemical gradients that the cell uses to drive work in the cell.
Most biologically important molecules are either too big (like glucose) or too charged (like ions) to pass through a membrane spontaneously. Transport proteins help these molecules to go across a membrane even if their chemical properties prevent them from diffusing spontaneously. The selective transport of molecules may or may not require the input of energy, like ATP, or rely on concentration gradients. The Amoeba Sisters have an excellent review video (Video 02-02) that explores the selective permeability of the membrane and how transporters regulate molecules going in and out of the cell.
Having control over which molecules are able to cross a given membrane is a key factor in how cells and organelles function. For example, the mitochondria’s capacity to generate ATP is almost entirely dependent on the formation of a concentration gradient (discussed in Video 02-02, above). Concentration gradients are used extensively by the cell to do work. They are analogous to a hydroelectric dam. A dam holds water in an upstream reservoir and controls the flow of water through turbines to create energy. In this analogy, the membrane is the dam, and specific ions are held at different concentrations on either side. As the ions naturally “flow” from the side of the membrane that has a high concentration to the side that has a low concentration, energy is released. The release of the molecules down the gradient is often used to “power” other cellular processes and thus is a key part in understanding advanced physiology. In this textbook, due to the metabolic pathways that we focus on, we mostly discuss the formation of proton gradients (i.e., H+ ions), but other electrochemical gradients are equally important to proper cellular function.
General Features of Biological Membranes
The Membrane Is Organized but Fluid
Lipids have a key role to play in the organization and fluidity of a membrane (a key concept). Proteins in the membrane, while also organized, may or may not be fluid, depending on whether they are anchored to internal or external structures. First, we will explore how we define fluidity in this context.
There are two kinds of fluidity in membranes that must be considered:
- lateral motion of lipids within a single leaflet of the bilayer and
- overall “stiffness” of the membrane.
Let’s look at each of these briefly in turn.
Lateral Motion of Lipids within a Single Leaflet Is Far More Favorable Than Movement between Leaflets of the Bilayer
The hydrophobic effect keeps the nonpolar portions of membrane lipids in the center of the membrane and is the reason membranes form spontaneously. As long as the nonpolar portions of the phospholipids stay in the interior of the membrane, their movement is not restricted (Figure 02-05). Within a membrane or bilayer, individual phospholipid molecules can spin rapidly on their axis and/or diffuse laterally within the plane of the bilayer. On the other hand, they very rarely flip-flop across the bilayer, as it would require the polar head group to pass into the nonpolar center of the membrane.
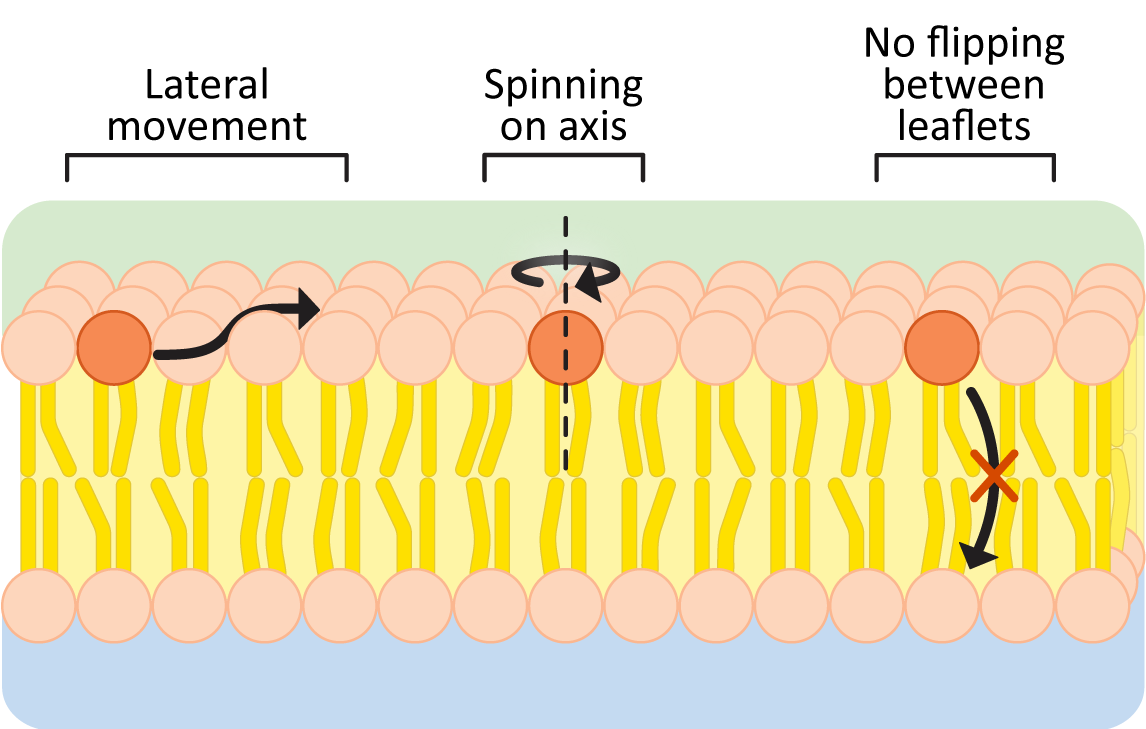
To further illustrate this point, here are some data to convince you:
- the frequency of lateral shifts of a phospholipid within the same layer is roughly 106/sec, but
- the frequency of flips from one leaflet to the other is closer to 10–5/sec.
Thus, flips between bilayer leaflets are about 1011 times less frequent than lateral movements within a phospholipid layer. Based on these numbers, an individual phospholipid would “flip” to the other leaflet about once every 28 hours on average. Thus, when we discuss membrane fluidity, we are generally referring to the lateral mobility of membrane components within a single leaflet of the membrane. It is also why we often refer to membranes as two-dimensional fluids.
Functional biological membranes require some movement of their lipids. The movement of lipids allows for rapid resealing of membranes in response to small holes or tears. However, too much fluidity can be damaging. A membrane that allows too much motion might have trouble keeping everything in its correct location. Think about a boiling pot of water, where it’s boiling so hard that water is spilling out everywhere. Membranes with too much fluidity may become holey or lose vital parts, which will make it challenging for the membrane to maintain its functionality.
Membrane Composition Also Determines How “Stiff” a Membrane Is
Membrane stiffness refers to how pliable or bendable the membrane is. It is a separate but equally important component of membrane fluidity, which is determined by the composition of the membranes. If membrane lipids pack too close together, the membrane will freeze in place, and the result will be a membrane that is too rigid to adapt as the cell moves and changes. On the other hand, a membrane that is too pliable will struggle to maintain the shapes required. Thus, the cell needs to manage the fluidity of its membranes in more ways than one. The cell manages this by controlling the precise lipid composition of the membrane. Remember that within each grouping of membrane lipids (phospholipids, glycolipids, sphingolipids) is a large family of similar molecules. By changing which phospholipids (or sphingolipids, etc.) and how much cholesterol is in the membrane, the cell can maintain its membranes within the correct range. That way they will be fluid enough to allow movement and bending but not so fluid that the cell struggles to control function. In Topic 2.2, we will look more deeply at how this fine balance is maintained by the cell in all kinds of environmental conditions.
General Features of Biological Membranes
The Membrane Is Asymmetric
Since membranes are exposed to different compartments and different environments on either side, it stands to reason that one would not expect the membrane, or its composition, to be identical on either side as well. This phenomenon is referred to as membrane asymmetry. There is always an inside that faces the cytoplasm (cytoplasmic side) and an outside facing the interior of an organelle or, in the case of the plasma membrane, the external environment (extracellular side). Both the lipids and the proteins play an important role in membrane asymmetry.
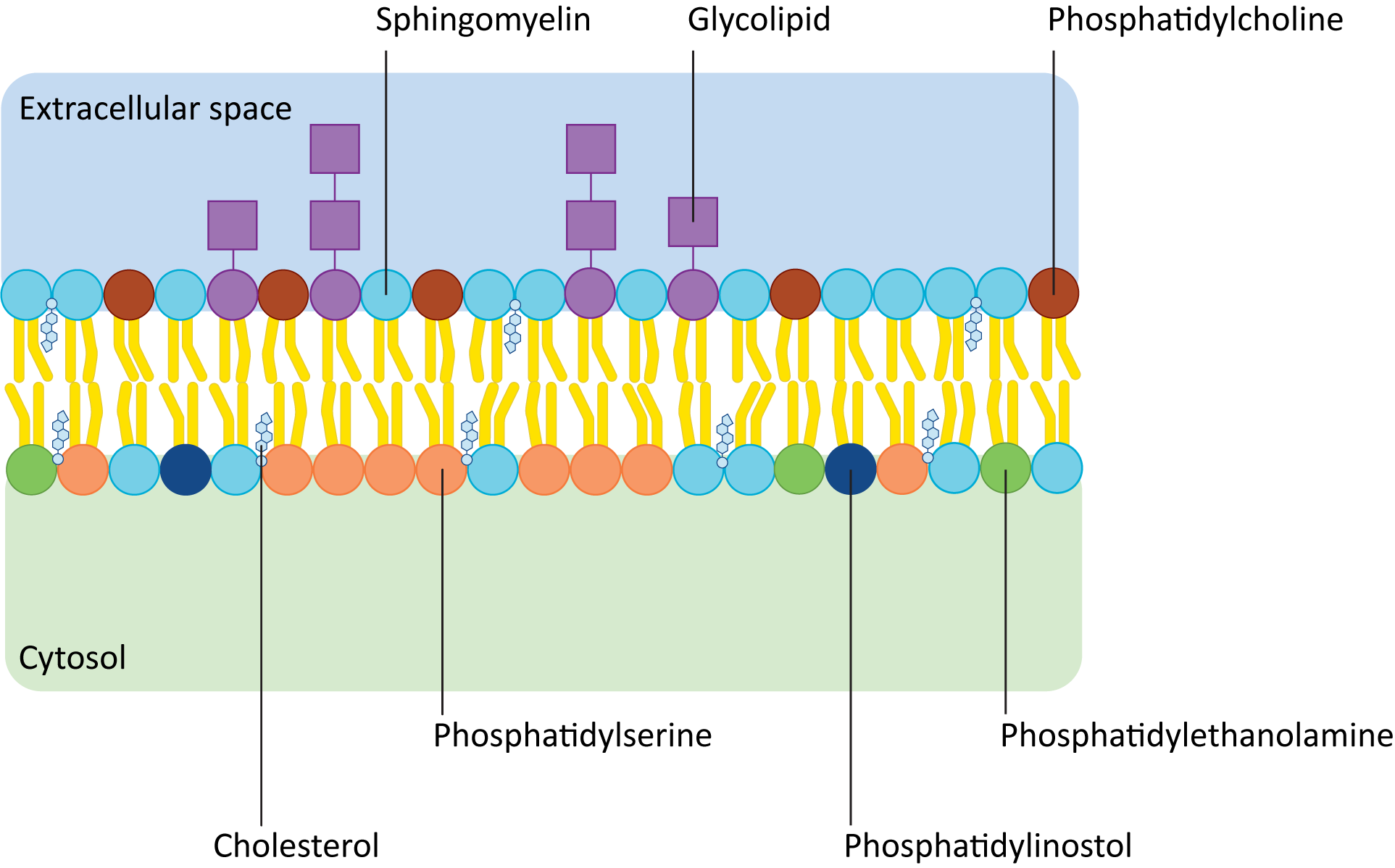
Figure 02-06 shows how the composition of lipids typically differs in the two leaflets of the plasma membrane. Remember that the term phospholipid refers to a whole family of lipids, so even though leaflets are made of phospholipids, the exact composition may not be the same. In addition, there are other kinds of lipids (e.g., sphingolipids and glycolipids) that might be enriched in one side of a membrane versus the other. For example, glycolipids are almost never found on the cytosolic side of the plasma membrane. Glycolipids face the cell exterior, as they play key roles in extracellular signaling and cell identity. Within the cell, organelle identity is also signaled by specific membrane lipids known as phosphatidylinositols, or PIPs for short. PIPs face the cytosolic side of the membrane and are used extensively by signaling and vesicle transport machinery.
The proteins in a membrane are also asymmetric. The parts that are sticking out of each side of the membrane will naturally be different to accommodate their specific functions. For example, a receptor protein will need to have the signal-binding site on the exterior surface of the membrane where the signaling molecule is present. Thus, each membrane protein needs to be inserted into the membrane in a very specific way to preserve proper function. Thus, the two sides of a membrane are not interchangeable, and the cell must keep them properly oriented at all times. We will discuss how this orientation is maintained in specific processes, like vesicle trafficking, in Chapter 4.
For Many Membranes, Carbohydrates Are Also Major Contributors to Membrane Asymmetry
At this stage, it is essential that we take a moment to point out the role of carbohydrates, the fourth major biological macromolecule. While they sometimes get overlooked in introductory cell biology courses—simply because there are so many other things to cover—they are absolutely essential to proper cellular function. The plasma membrane of animal cells is usually completely covered with carbohydrates—most commonly attached to proteins but also to lipids. This coating (called the glycocalyx) plays a key role in cellular identity and signaling. Plants and fungi have so many carbohydrates surrounding them that they form a structure known as a cell wall. We will take a moment at the end of this chapter to explore both the extracellular matrix and plant cell wall in a little more detail.
The Origin of Membrane Asymmetry
While it’s easy to understand the need for membrane asymmetry, it is much more complex to understand how asymmetry is created by the cell in the first place. In order to understand that, we must explore how membrane lipids are made by the cell.
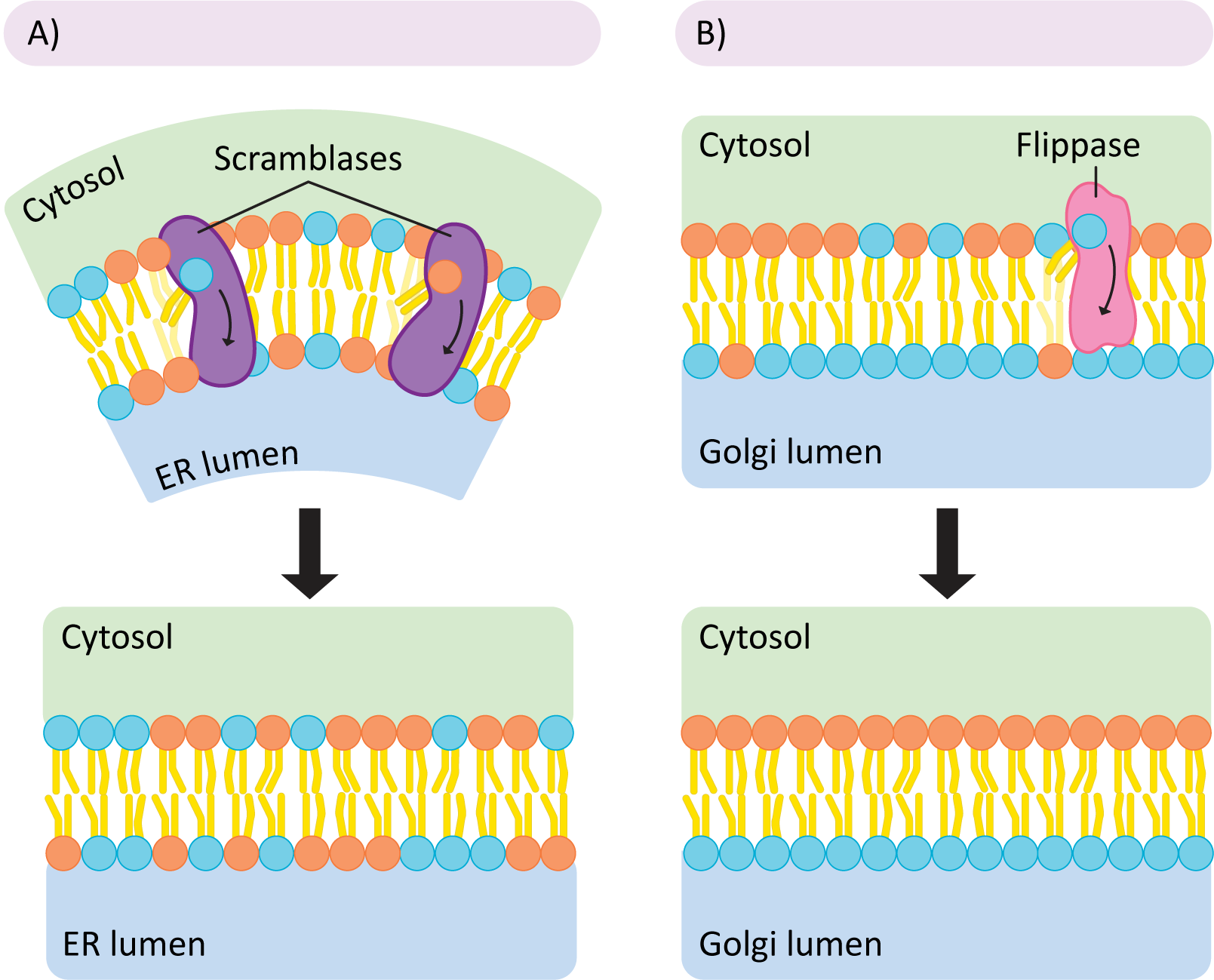
All membrane lipids are first synthesized in the cytosol and then inserted into the cytosolic face of the smooth ER. This means that all new lipids are being added to a single leaflet of the ER membrane. As you can well imagine, this causes pressure on the ER membrane, as one leaflet is growing while the other is not. If left this way, the pressure would eventually cause a potentially catastrophic rearrangement as the membrane returns to a more thermodynamically favorable conformation. To avoid this kind of event, the cell uses proteins known as scramblases to rebalance the lipids in each leaflet (Figure 02-07A). This is done nonselectively—the scramblase simply equilibrates the phospholipids on either side of the membrane regardless of type. Later, in other regions of the ER, but more commonly in the Golgi, the membrane is organized more precisely. Enzymes called flippases move specific phospholipids unidirectionally from the exterior leaflet to the interior one so that membrane asymmetry is properly established, and the lipids are placed where they need to be for function. Both enzymes use a similar mechanism—the enzymes are big enough to span the membrane, and the polar head group is slid through a slot in the enzyme so it can cross the nonpolar part of the bilayer (the tails stick out and are left free to the environment; see Figure 02-07). Thus, they can overcome the energetically unfavorable parts of the reaction and flip lipids from one leaflet to another.
Topic 2.2: Maintaining Fluidity in the Membrane
Learning Goals
- Describe how the lipid composition can influence membrane fluidity.
- Explain how membrane fluidity is maintained under different environmental conditions, like temperature.
- Explain how fluorescence recovery after photobleaching (FRAP) works, and interpret outputs from these experiments.
In the first topic of this chapter, we focused on the major characteristics of biological membranes as a whole, and how the chemistry of the membrane can contribute to its properties. Here we’ll go into more depth about how the fluidity of a given membrane is the product of its precise lipid composition. We will also learn about an experimental technique known as fluorescence recovery after photobleaching (FRAP), which allows us to assess the mobility of molecules within the membrane.
in depth
Membranes Are Organized and Fluid
When we think of the term membrane fluidity, the first thought might be that this simply indicates how quickly any given molecule can move from point A to point B within the membrane. However, this is only one aspect of membrane fluidity. As we mentioned in the previous topic, the stiffness of the membrane must also be considered.
Membrane stiffness describes how easily the membrane bends (compared to how easily or quickly the molecules within the membrane can move around). Membranes vary in how stiff they are and how they respond to their shape being distorted. For example, red blood cells are elastic, which means they can be distorted but bounce back to their original shape. On the other hand, white blood cells are much more rigid and don’t deform as easily. As an analogy, a red blood cell is like a plastic bag, and a white blood cell is more like a cardboard box. This is the result in differences in both the lipids and proteins that are associated with each cell’s plasma membrane. The composition of the membrane is key for the cell’s capacity to function in its environment.
Membrane Fluidity Is Heavily Influenced by the Environment and Must Be Maintained at a Constant Level
Organisms live in all kinds of environments, from the hottest regions of the Sahara to the coldest regions of the Arctic and everything in between. They also live in regions of extreme pressure, like the deep sea, which can impact both the freezing and boiling point of water. In all of these extreme environments, cells must control the chemistry of their membranes so that they remain fluid and functional. For example, membranes must not freeze when exposed to low temperatures (or high pressure) or lose their integrity at higher temperatures. Any organism that lacks a mechanism to control the fluidity of its membranes will die if/when environmental conditions change.
There are two major approaches that organisms take to controlling the fluidity of their membranes:
Approaches to Membrane Fluidity |
|||
|---|---|---|---|
Endotherms |
Ectotherms |
||
Pros |
Cons |
Pros |
Cons |
|
|
|
|
This section naturally focuses on exotherm membranes, whose membrane composition changes more in response to the environment. One good example of membrane composition being used as an environmental adaptation is cold-hardiness in plants. As the temperature lowers in the fall and winter, membrane lipids become increasingly unsaturated. This lowers the melting/freezing point of the membrane, which helps it maintain flexibility, and retains the mobility of the membrane components. This also explains why organisms might be fragile with respect to sudden, unexpected changes in temperature. In the example of the plant, a rapid drop in temperature at the wrong time (like a cold snap in early autumn) could result in the death of the plant, as they wouldn’t have the time required to synthesize new membrane lipids (and other antifreeze proteins, which help avoid freezing).
Membranes That Are Unable to Adapt Chemically Are Susceptible to Failure in Nonoptimal Temperatures
The capacity of a membrane to remain fluid at any given temperature is primarily dependent on its capacity to maintain just the “right” amount of entropy in its phospholipid tails. If they pack too close together, they will freeze, and if they have too much motional freedom, they will fail in other ways.
When a membrane (or anything else) freezes, what really happens is that as the temperature lowers, the kinetic energy of the molecules is also lower, so they become sluggish and do not move around as freely. At some point, the kinetic energy of the phospholipids is so low that they get drawn into the transient induced dipole–induced dipole interactions that continue to happen between neighbors and can no longer break away. Eventually, the phospholipids will get “trapped” by the intermolecular forces being exerted on them by their neighbors, and the lipids will freeze in place. In chemistry terms we say that the membrane is in “gel phase.” You can see the result of this any time you pull a piece of frozen meat from the freezer. It is quite stiff and inflexible compared to its room-temperature counterpart.
At the other extreme, when the temperature is too high for the membrane, the molecules have quite a lot of kinetic energy. If the kinetic energy is too high, then the intermolecular forces that hold the membrane together are too easily broken, and molecules can break free all together. When this occurs, important complexes will come apart, ions and other molecules may be able to slip through the chaotic membrane lipids, and the membrane itself could just fall apart.
Maintaining Fluidity—Membrane Composition
Cells have three different ways that they can modulate in order to influence the fluidity of a given membrane.
-
Change the Degree of Unsaturation of the Hydrocarbon Chains
Increasing the unsaturation of the lipid fatty acid tails increases fluidity of the membrane. Unsaturated lipids contain one or more double bonds between carbons in the hydrocarbon chain (compared to saturated lipids that have only single bonds in their hydrocarbon chains). The double bonds change the shape of the tail because they create bends, or “kinks,” in the hydrocarbon chain (Figure 02-08). A kinked tail will not be able to pack as tightly with its neighbors. Imagine that on a packed subway everyone stands wide-legged, with their elbows sticking out. The net effect would be that each person would take up more space, and fewer people would be able to get into each car. The net effect of unsaturation is essentially the same—less tight packing, which effectively reduces the number of induced dipole–induced dipole bonds between lipids in the membrane. The result is that the lipids are able to maintain their mobility even at the reduced kinetic energies of lower temperature.
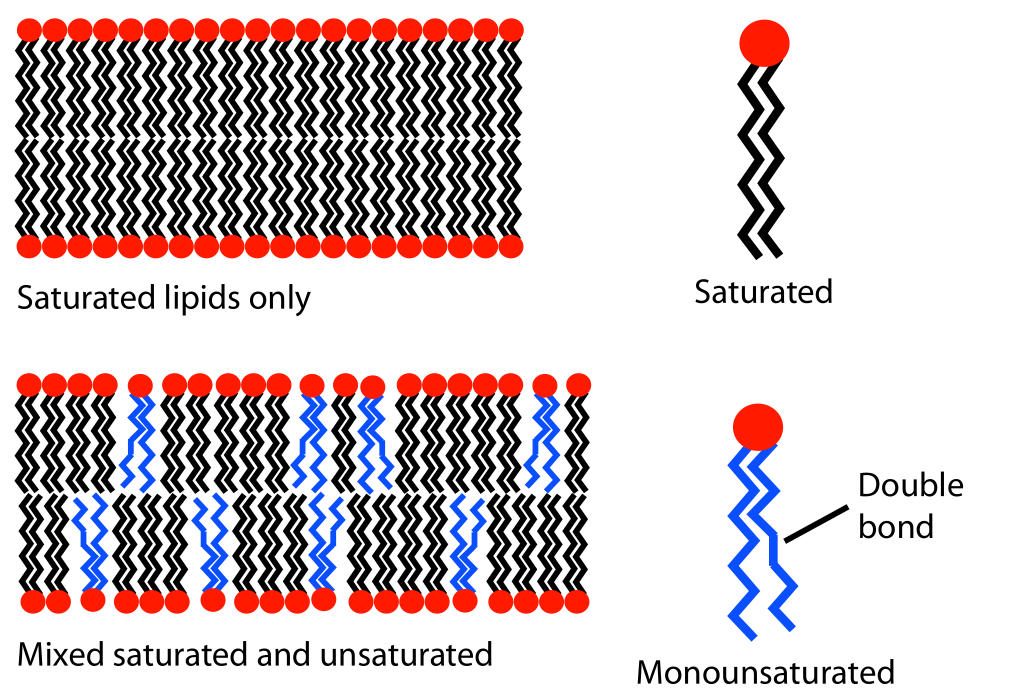
Figure 02-08: Depiction of how unsaturated phospholipids can impact the packing of lipids in a bilayer. Lipid unsaturation effect.svg by MDougM. This image is in the public domain. -
Change the Number of Carbon Atoms in the Fatty Acid Tails
Shortening the fatty acid tails of a phospholipid will have a similar effect to using unsaturated fatty acid tails in that it decreases the number of transient bonds that can form between neighbor phospholipids. Longer hydrocarbon chains have more atoms in them; thus, there is the potential for more bonds to form. By shortening the hydrocarbon chains, the cell reduces the total number of atoms that can get involved in bonding at once, thus reducing the amount of energy that can be used to hold neighbor phospholipids together. Therefore, membrane lipids with shorter hydrocarbon chains will tend to have more mobility at lower temperatures. Conversely, regions of membranes with longer fatty acid tails can be used to reduce overall fluidity in that region and to hold specific proteins within that region.
-
Change the Sterol Content of the Membrane
Cholesterol is a type of sterol (ringed lipid) that sits in the spaces between fatty acid chains (Figure 02-09). At first glance, based on what we have previously learned about creating space to reduce bonding, one would expect that this would be beneficial at low temperatures, as it would reduce the interactions between the fatty acid tails of the phospholipids. This is exactly true. At lower temperatures, increasing the amount of cholesterol will also increase membrane fluidity. But in this case, the story doesn’t end there.
Paradoxically, cholesterol also has an effect at higher temperatures. However, in this case, increasing cholesterol content will reduce the fluidity of the membrane. It does this by filling the space in between lipids that is created when lipids have high kinetic energy. They also have a stiff ring structure, which also contributes to the stiffness of the membrane. Cholesterol at high temperatures increases the number of intermolecular forces between neighbors, which is important to stabilize the lipids in the membrane.
Think of cholesterol as a fluidity buffer. It helps keep the membrane within a specific range of fluidity so that it doesn’t get too rigid or too fluid. Modulating the cholesterol can help control fluidity at many temperatures, which makes it an important membrane lipid in all environments. (Note: Cholesterol is unique to animals. However, sterols are an important component of all eukaryotic membranes. Even though plants do not make cholesterol, they do use other phytosterols for the same purpose.)
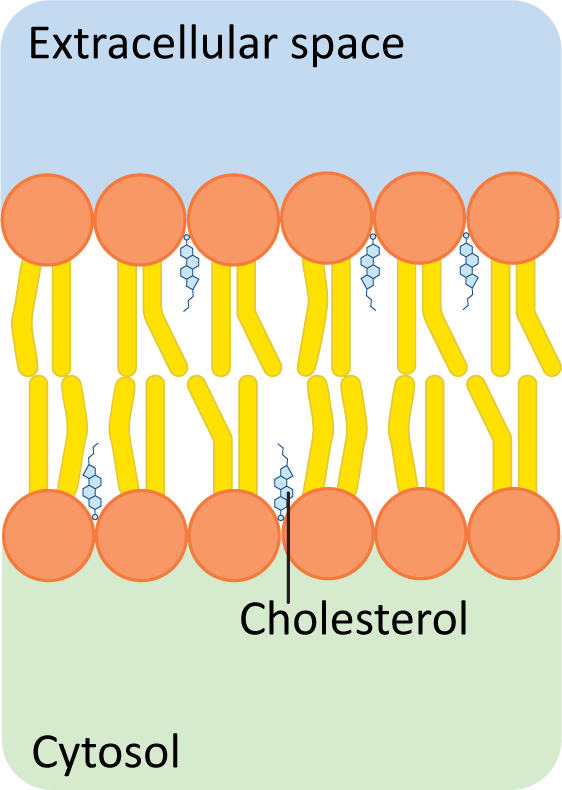
Figure 02-09: Cholesterol fits between the other lipids, inside the hydrophobic region of a phospholipid bilayer. Because cholesterol has a very small hydroxyl group as its only polar component, the majority of the molecule sits in the hydrophobic tail region. This image was created by Heather Ng-Cornish and is shared under a CC BY-SA 4.0 license.
“Real” Membranes Contain Regions with Varying Fluidity
While it might be tempting to think of the cell membrane as a homogenous thing that is the same across the entire cell, there is significant evidence that this is not true. Both the lipid and protein components of any given membrane will be different in different regions, as different functions will need to occur. For example, lipid rafts are distinct regions of the membrane that contain a higher concentration of sphingolipids with longer fatty acid tails and cholesterol (Figure 02-09). The increased length of the sphingolipid tails in that region helps hold the lipids together and also increases the thickness of the membrane in these regions. Cholesterol also plays an important role in the structural integrity of the lipid raft. Indeed, when cholesterol is removed experimentally lipid rafts will fall apart. However the exact nature of its role is complex. It is said to act as both a “glue” that holds the raft together, and also as a buffer to stop the raft from becoming overly rigid. Specific proteins are preferentially drawn to these regions based on their chemical features (e.g., longer membrane-crossing domains). The specific clustering of proteins in lipid rafts can be used to keep proteins together that all work toward the same function (e.g., signaling or vesicle trafficking). A diagram of a lipid raft is shown in Figure 02-10.
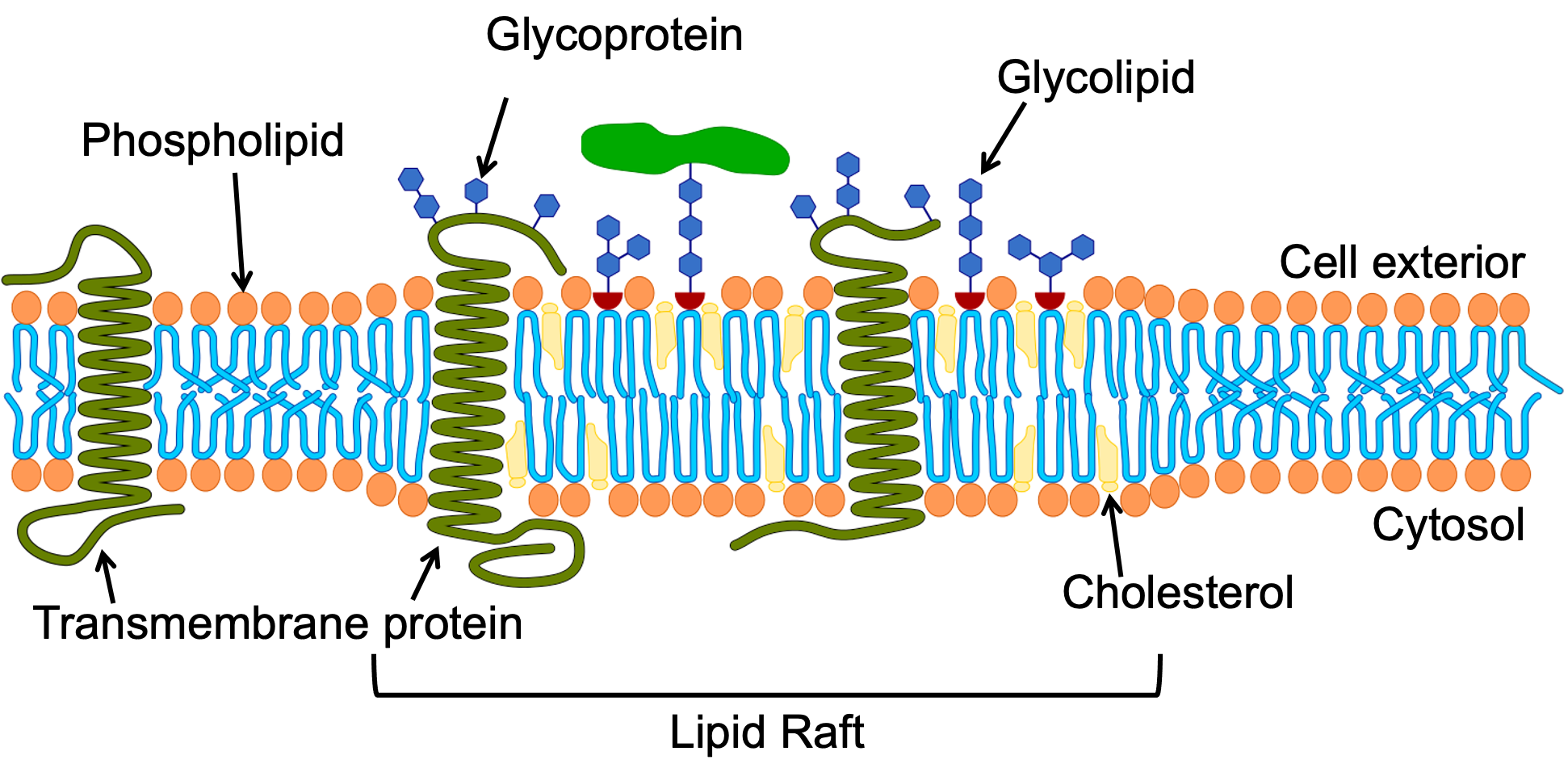
Studying Cells: Fluorescence Recovery after Photobleaching (a.k.a. FRAP)
One of the experimental tools that is used to examine how cellular components move is a fluorescence microscopy technique known as FRAP. While it can be used to study any number of cellular functions that involve movement, in this case we will focus on how it can be used to track movement within a membrane.
In this technique, scientists fluorescently label a specific membrane component (usually a lipid or protein). Then the high-powered laser of a confocal microscope is used to photobleach the fluorescent tag, meaning that the fluorescence of the tag is extinguished without damaging the protein or lipid to which it is attached. A small, specific area of the cell is bleached, and then we track how long it takes for unbleached molecules to return to the bleached area. There is an excellent explanation of this in Video 02-03.
The video also shows us the kinds of results we can expect from these experiments. To summarize, graphs are generated from these data, with the fluorescence intensity in your region of interest on the Y-axis and time on the X-axis. Based on the amount that the fluorescence “recovers”—meaning the fluorescent intensity in that region increases again—researchers can compare how mobile a particular lipid or protein is within the membrane. We call this graph a recovery curve, as it shows how the fluorescence “recovers” in the region of interest over time.
Topic 2.3: Structure and Function of Membrane Proteins
Learning Goals
- Explain how the primary sequence and the environment of a protein influence its final 3D structure, specifically with respect to the different types of intermolecular forces that it will form with itself and its environment.
- Distinguish between integral and peripheral proteins with regard to their solubility properties, structure, and manner of attachment to membranes.
- Describe the different roles of membrane proteins within a biological membrane.
- Describe the structure and use of plasma membrane–adjacent structures like the animal cell extracellular matrix and the plant cell wall.
Earlier in this chapter, we learned that a biological membrane is different from a phospholipid bilayer. A biological membrane also has proteins embedded in it that will change its properties and add functions. In this topic, we will first review the formation of proteins from amino acids, then discuss the characteristics of membrane proteins. We will also examine the unique properties of the plasma membrane as the barrier that separates the cellular contents from the outside world. We end with a brief discussion of what’s on the outside of a cell—namely, the extracellular matrix (animals) or a cell wall (plants, algae, and fungi).
The protein content of a membrane can range from ~25% (in the case of the myelin sheath cells of neurons) to nearly 80% (for the mitochondrial inner membrane), with more typical membranes consisting of protein and lipids in an approximately equal ratio (50:50) by weight.
Because of the associated proteins, the thickness of biological membranes is almost always thicker than that of a simple lipid bilayer. Biological membranes are typically 6.5 to 10 nm thick. A lipid bilayer without proteins is about 5.5 nm thick.
Brief Review of Amino Acids and the Chemistry of Protein Folding
We assume you have preexisting knowledge about amino acids and proteins, so we are approaching this as a review. As always, we encourage you to explore the resources in the introduction if you need a refresher.
There are 20 different types of amino acids that are used to form proteins. Each amino acid contains an amino group (NH3+) and a carboxylic acid group (COO–). In between is a carbon atom connected to a variable chemical structure called the R group or side chain. To make a protein, the amino group from one amino acid will covalently bond with the carboxylic acid group of another amino acid to create a peptide bond. This creates a repeating N–C–C pattern when the amino acids are strung together. This repeating N–C–C is called the protein backbone.
The sequence of the amino acids in a polypeptide is based on the sequence of codons that are read from the mRNA by the ribosome during translation. We call the order of the amino acids in the polypeptide chain the primary structure or primary sequence.
The R groups are what give each amino acid its distinct chemical properties. Some R groups are large, and some are small; some are negatively charged at physiological pH, and some are positively charged. It’s worth taking some time to look at the chemical structure of the R groups and start investigating how these chemical groups impact protein structure/function.
The order of the amino acids in the primary structure plays a crucial role in determining folding and function of a protein. A given protein will fold in a very specific way depending on the molecular interactions of the amino acids in its primary structure. A variety of intermolecular forces (i.e., hydrogen bonds, ionic bonds, and induced dipole–induced dipole/van der Waal forces) contribute to the final 3D fold of the protein. Proteins will, many times, spontaneously fold into the shape that is the most stable and requires the least amount of energy to maintain. Other times, chaperone proteins will help facilitate the proper folding of a protein. We will learn about chaperones in more detail in Chapter 4.
When discussing protein folding, we split the different interactions into four categories, which we call the “levels” of protein folding. It’s important to remember that these do not happen sequentially. Instead, they happen more or less all at once in different parts of the protein as it folds into its final 3D shape. These levels are as follows:
- Primary structure forms when amino acids are covalently bound together by peptide bonds. The order of the amino acids is the key feature of the primary structure.
- Secondary structure forms when the backbone interacts with itself via hydrogen bonds. It forms a repeating local pattern, commonly with nearby amino acids. Examples of this structure include alpha helices and beta sheets.
- Tertiary structure forms when R groups get involved. They interact with either other R groups or the backbone, usually via intermolecular forces like the ones mentioned above.
- Quaternary structure forms when different polypeptide chains come together to form a protein (or protein complex). All of the same intermolecular forces get used as we would expect in tertiary structure.
Disulfide bridges are the only other covalent bond used in folding aside from the peptide bond that creates the primary structure. Disulfide bridges form between two cysteine residues. They can play a role in tertiary or quaternary structure but are far less common than the other bonds. To compare, there are usually only a handful of disulfide bridges in a protein, if any, whereas there may be hundreds, or even thousands, of hydrogen bonds. As a result, we tend to discuss disulfide bridges much less often, but that doesn’t mean that they should be forgotten.
The chemical environment (especially pH) also plays a very important role in protein folding. A soluble protein in the cytosol will experience different intermolecular forces than a protein that is embedded in a membrane. In each case, the protein will respond to its folding environment. Unlike the soluble proteins of the cytosol, many membrane proteins must be able to embed themselves in the lipid bilayer, including the nonpolar tail region. This requirement impacts how the protein folds and which amino acids of its primary structure are expected to be on the exterior in each region.
Integral and Peripheral Membrane Proteins
Proteins can be associated with the membrane in multiple ways. At the most fundamental level, proteins may
- exist solely on the surface of a membrane, living peripherally on one side or the other, or
- extend into the nonpolar tail region of the membrane in some way so that they are integrated into the membrane itself.
This may sound like a very simple thing to differentiate: Do they exist on the surface or extend into the membrane itself? However, it isn’t always as easy as it sounds. For example, how do we categorize a protein that sits on the surface but also has a covalently linked lipid tail that extends into the membrane? (Answer: We consider it to be integral.)
Since proteins in membranes are too small for us to see, the categorization of proteins has historically been based on experimental evidence. So if we include the experimental evidence in our definition, we come up with the following:
- Integral membrane proteins are proteins that cannot be removed from the membrane without destroying the membrane completely. Usually, this requires the use of strong detergents, which disrupt the structure of the membrane so that proteins can be removed. Only proteins that extend into the membrane in some way will require this level of disruption to extract.
- On the other hand, peripheral proteins are much easier to remove. Usually, a simple ionic salt wash is enough to dislodge these proteins from the membrane, as the intermolecular forces are not as strong and can be more easily disrupted.
| Integral vs. Peripheral Membrane Proteins | ||
|---|---|---|
| Criterion | Integral proteins | Peripheral proteins |
| Requirements for removal from the membrane | Strong detergents or organic solvents used to break up hydrophobic associations | Mild detergents, high salt concentrations, and/or metal chelating agents used to remove |
| Association with lipids after extraction | Usually remain partially associated with lipids after removal | Usually completely free of lipids once removed |
| Solubility after extraction | Usually insoluble or clumped together in neutral aqueous solutions (e.g., water) | Soluble and dispersed in neutral aqueous solutions |
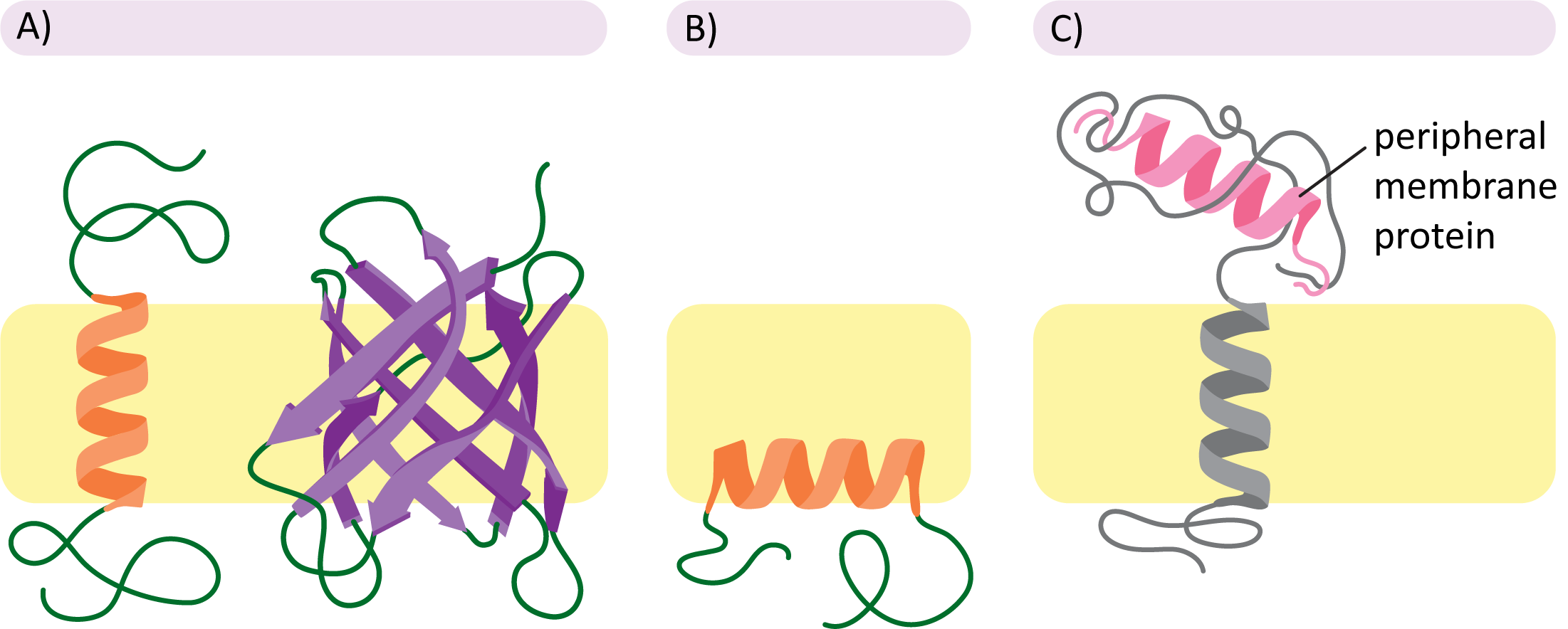
Figure 02-11 also helps illustrate the different subcategories of membrane proteins:
- Transmembrane proteins (Panel A): These integral membrane proteins cross the entire membrane and stick out on either side into the cytosol. These proteins are held in place primarily through the hydrophobic effect. More on that in the section below.
- Monolayer-associated proteins (Panel B): This is also a type of integral membrane protein. Although they are not considered to be very common, they still occur rarely. In this case, the protein is held in place by an alpha helix that is amphipathic (i.e., nonpolar on one side only). We will not be discussing this type further.
- Peripheral membrane proteins (Panel C): These proteins are most commonly (but not always) attached to the membrane through associations with integral membrane proteins. These linkages are most often via ionic or hydrogen bonding or some combination of several types of intermolecular forces. The association of peripheral proteins with one side of the membrane or the other further contributes to the asymmetry of the membrane.
Transmembrane Proteins Require Specific Secondary Structure to Be Able to Pass through the Membrane
One of the biggest challenges faced when proteins must pass through a membrane is how to deal with their backbones. Unlike the composition of R groups, the backbone of the polypeptide chain is always polar. The H, N, and O atoms that make up the peptide bond are electronegative and capable of forming hydrogen bonds. As such, they face a thermodynamic challenge when required to pass through the hydrophobic center of the lipid bilayer. So how is this addressed in the cell?
The answer lies in their secondary structure. As you may recall, secondary structures are defined as local, repeating structures that are formed via hydrogen bonding of backbone atoms to other backbone atoms. These repeating structures usually have a twofold effect:
- they allow the protein backbone to form hydrogen bonds with itself, which is thermodynamically stable, and
- they usually push the R groups outward, where they are available to interact directly with the environment. This leaves the protein backbone in the center, sequestered away from the nonpolar lipid environment.
The two secondary structures we mentioned earlier are the alpha helix and the beta sheet. Both of these structures are very commonly found in transmembrane proteins. However, the details of how they form are naturally going to be different.
Alpha Helices in Transmembrane Proteins
Many transmembrane proteins have one or more alpha helices in their transmembrane domains. These alpha helices are stretches of about 20+ nonpolar amino acids (depending on the width of the membrane). Remember that in the alpha-helical arrangement, the amino acid R groups extend outward, and the backbone is in the center of the helix. By forming an alpha helix, the nonpolar R groups will shield the polar polypeptide backbone from the nonpolar environment in the center of a lipid bilayer, thus creating thermodynamic stability. Figure 02-12 shows an alpha helix from the side, embedded in a membrane. You can see the backbone structure represented by the purple ribbon winding up through the membrane. Hydrogen bonds between the atoms of the backbone (shown as dashed lines between the ribbon) hold the shape of the helix. The R groups of each amino acid in the polypeptide chain extend outward from the polypeptide backbone to interact with the tails of the phospholipids.
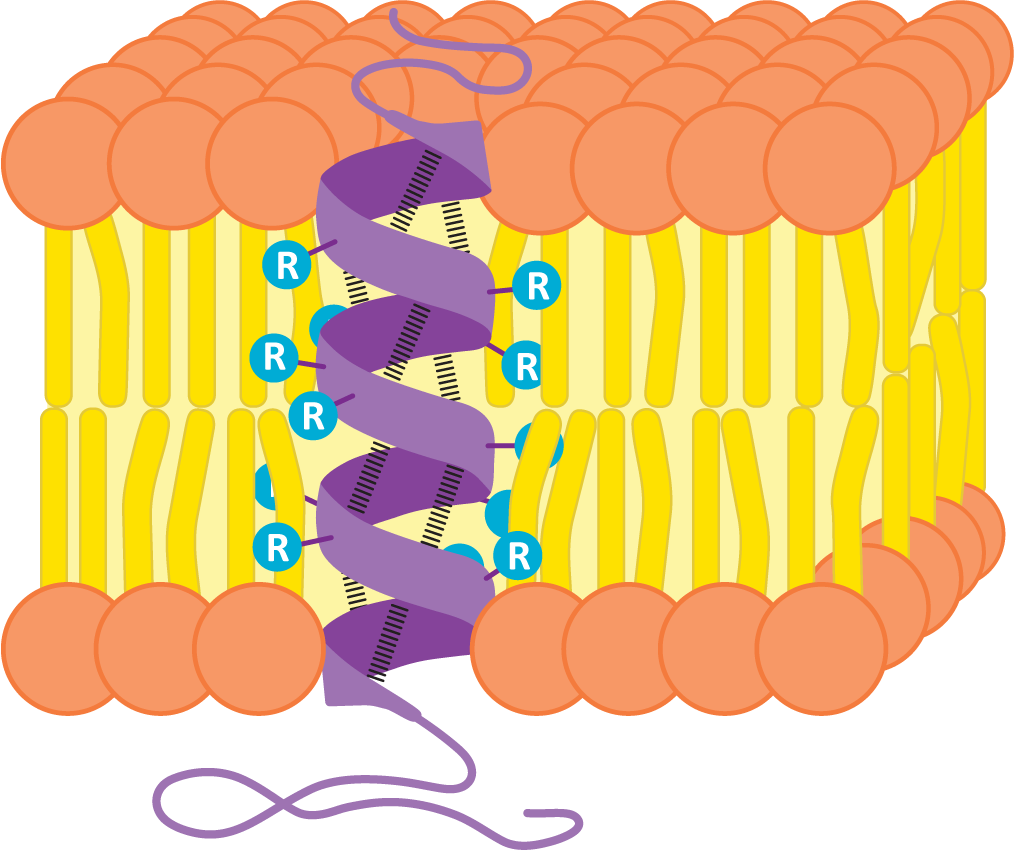
Since transmembrane proteins must be able to exist in both the nonpolar center of the membrane and the aqueous parts of the cell, it stands to reason that these proteins are amphipathic like other components of the biological membranes. The R groups of amino acids within the nonpolar portion of the membrane are nonpolar, whereas in other regions that interact with the aqueous environments, the amino acids will primarily be polar.
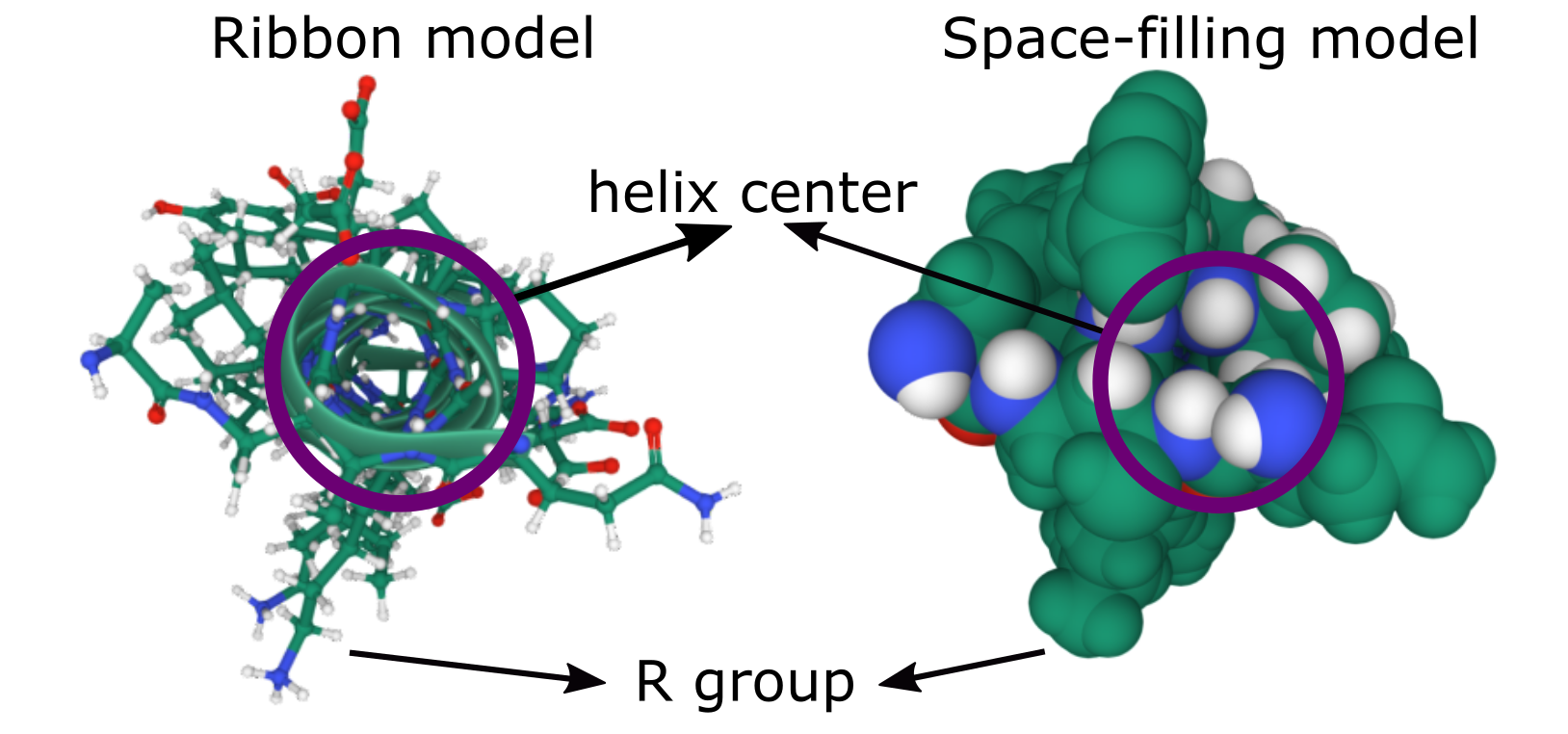
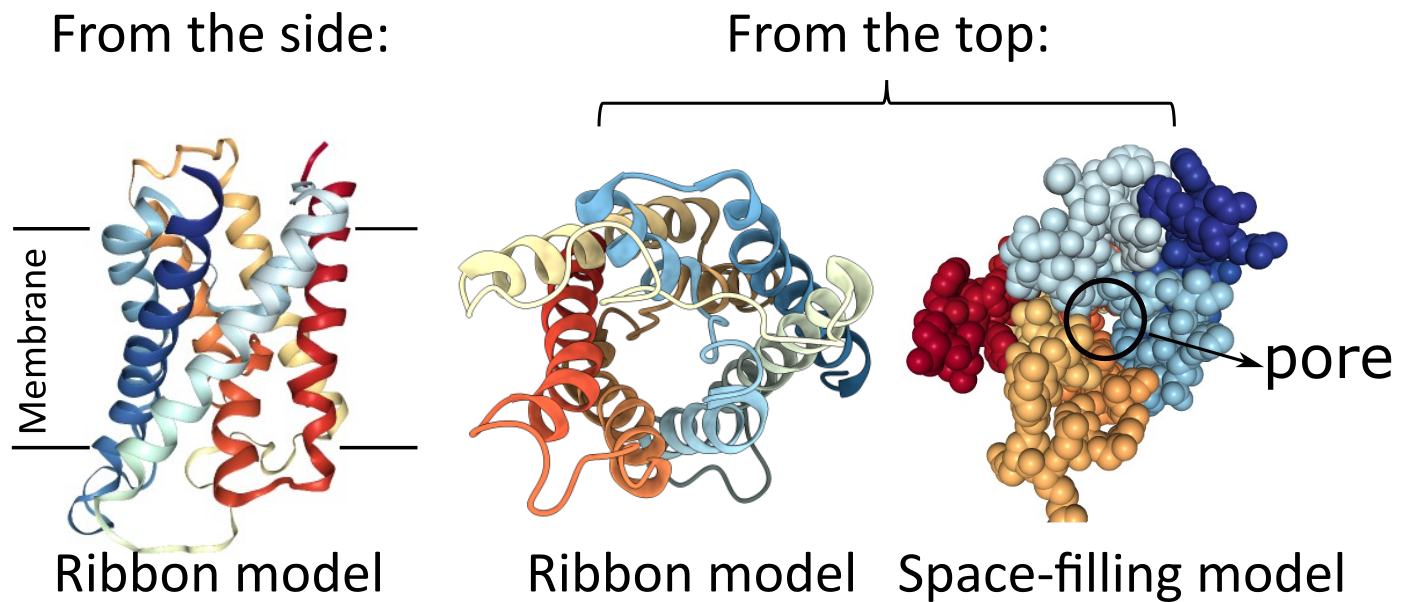
It is important to note that a single alpha helix does not form a channel, so it cannot allow anything to pass through it. The molecules of the backbone completely fill the space inside the helix (Figure 02-13). Alpha helices can be used to form the pores and channels that control the entry/exit of many molecules in/out of the cell. Because a single helix cannot act as a channel, several membrane-spanning alpha helices must instead cluster in a roughly circular arrangement through the membrane.
Aquaporin is an excellent example of a transmembrane protein that is made of several alpha helices, which are used to create a central channel (Figure 02-14). Aquaporin is nonselective and allows water and other small solutes to pass through the membrane. Note that the amino acids lining the interior of the channel will need to be polar and possibly charged in order to interact with the water and solutes that are expected to pass through. On the other hand, the exterior of aquaporin will have a large strip that will directly interact with the phospholipid tails of the membrane; thus, those regions will need to be nonpolar.
Beta Barrels in Transmembrane Proteins
While a single alpha helix is stable enough to be used as the sole membrane-spanning structure for a transmembrane protein, a beta sheet is not as well designed for this purpose. The edges of the sheet will have exposed backbone molecules that will not easily interact with the nonpolar portion of the bilayer. However, a beta sheet can circularize itself by hydrogen bonding the ends of the sheet together (Figure 02-15). This means that the backbone’s bonding requirements are met within the nonpolar region of the membrane, and we once again have an ideal scenario for a thermodynamically stable structure that can pass through a membrane. We call these circularized beta sheet structures beta barrels.
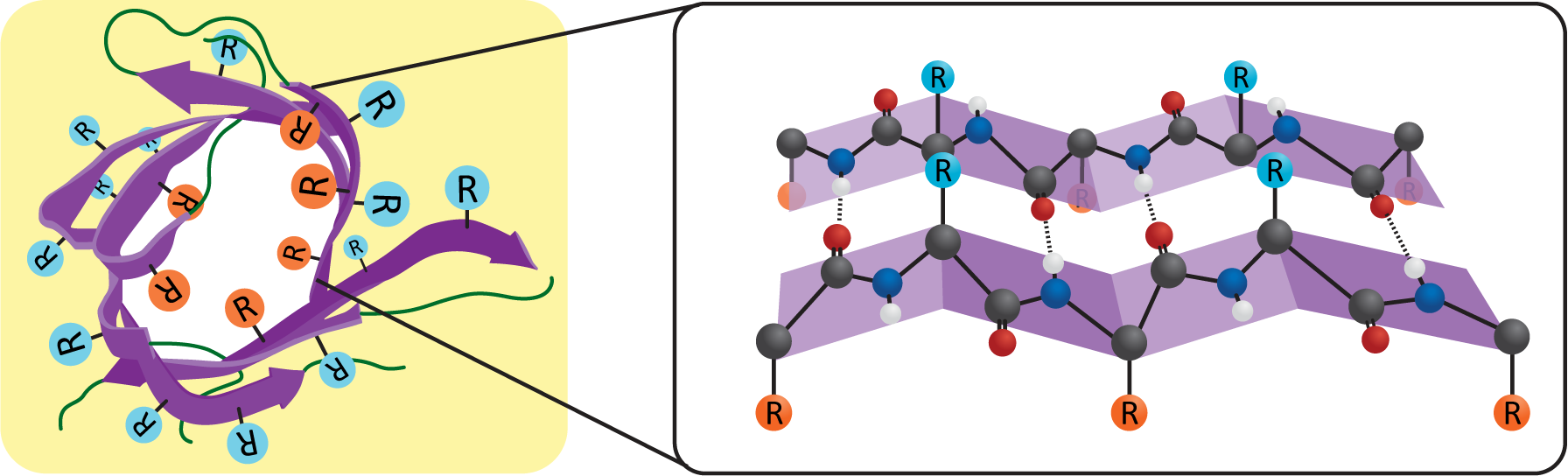
Like the alpha helix, beta sheets and beta barrels require a specific arrangement of their R groups in order to form. Unlike the alpha helix, the R groups stick out perpendicular to the face of the beta sheet in an alternating pattern (Figure 02-15). This means that if we were to unfold the beta barrel and examine the primary sequence, we’d likely see a series of amino acids with alternating properties (i.e., nonpolar, polar, nonpolar, etc.) because one amino acid with an R group facing the lipid environment would be next to an amino acid with an R group facing the aqueous pore environment.
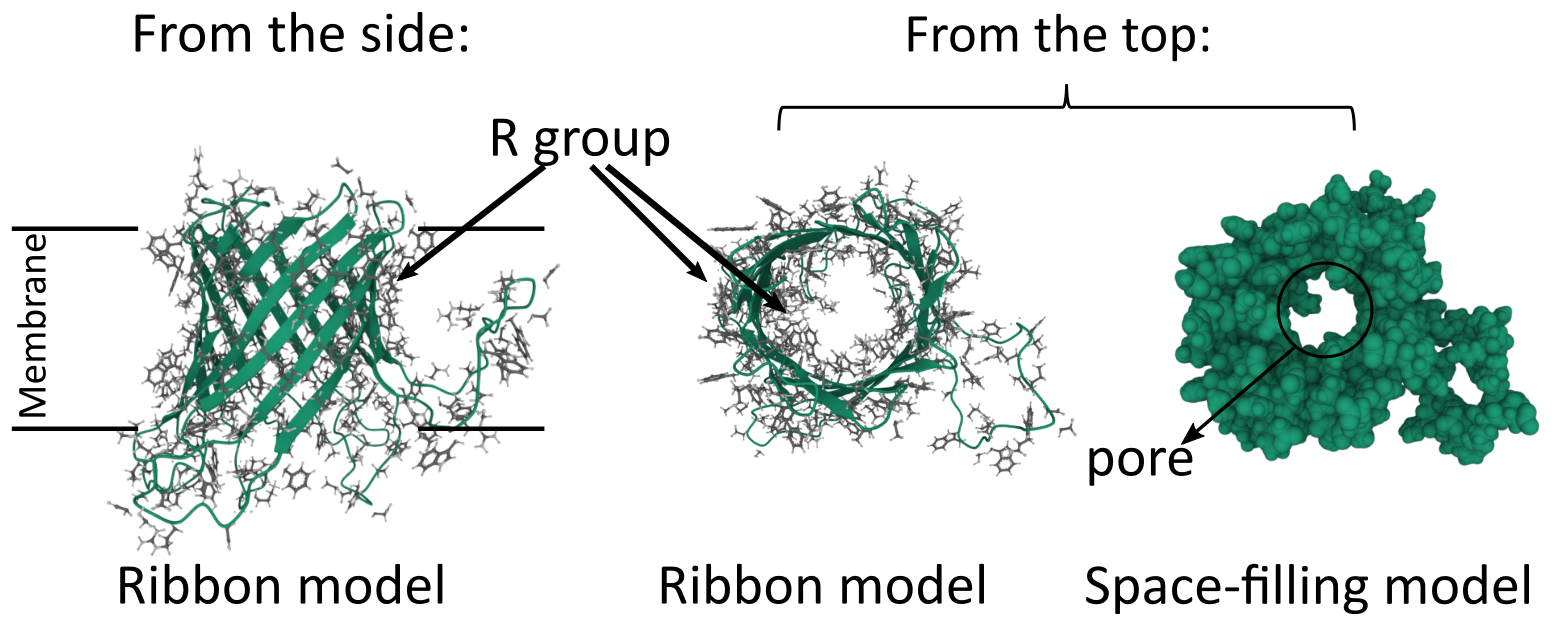
We can see this in action by examining the beta barrel structure of a real bacterial protein called outer membrane protein G (Figure 02-16). Once again, we see that this particular secondary structure, the beta barrel, precisely suits the function of the protein as a transporter. This protein is used to take up large carbohydrates. Thus, the outside of the beta barrel must be able to interact with the lipid environment and help hold the protein in the membrane, whereas the inside of the barrel must create enough space to allow specific molecules to pass through the membrane. In addition, the amino acids of the center channel must be polar as well to interact with the carbohydrates that are transported. In summary, the properties of the amino acids must match the chemical environment where they reside and function.
Function of Membrane Proteins
Membrane proteins carry out many different functions in the cell. It is important to remember that membranes in the cell have different sets of proteins in them, as they each must carry different functions. As such, the protein composition of the endoplasmic reticulum (ER) membrane is different from, say, the plasma membrane.
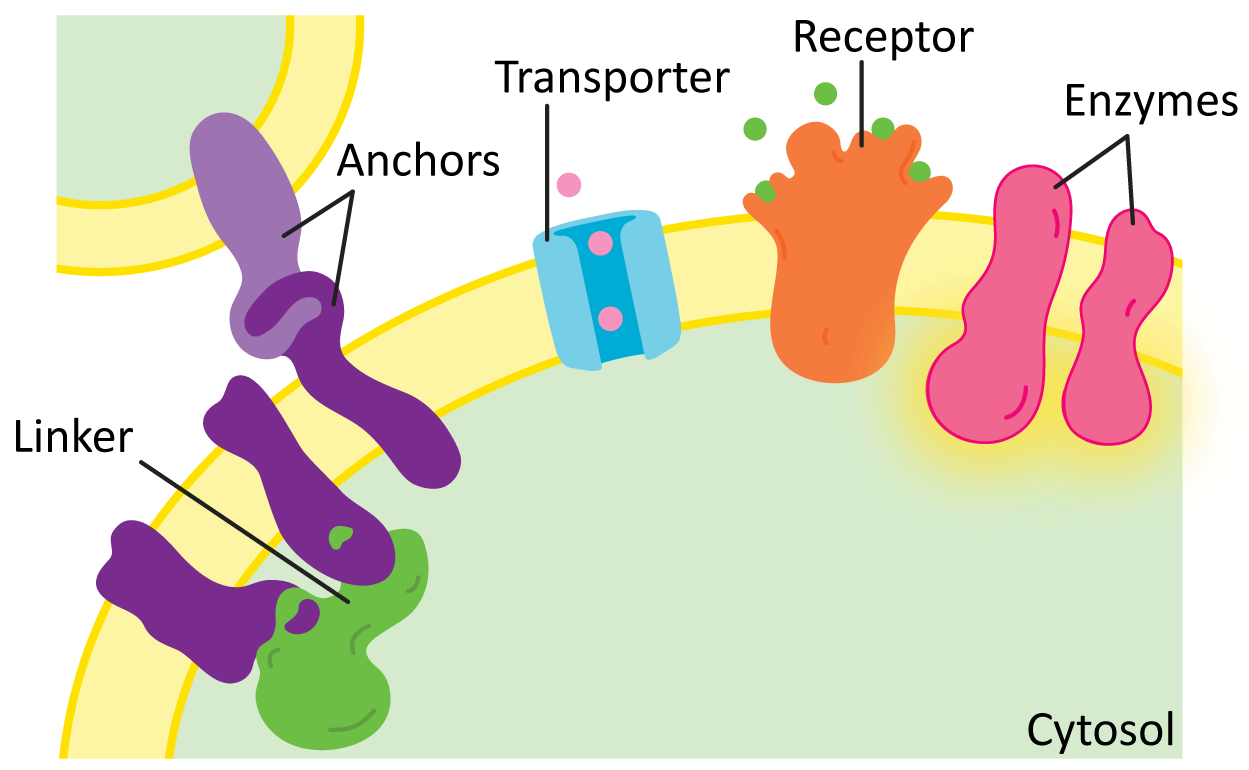
While there are many different functions for membrane proteins, for the most part they’ll fall into one of the following categories (Figure 02-17):
- Structural proteins, such as linkers and anchors—Anchors help attach the membrane to organelles, the extracellular matrix, or even other cells, whereas linkers help connect several proteins in the membrane and can help provide shape. We will see many examples of structural proteins as we discuss cellular function.
- Transporters—These proteins mediate the transport of different types of molecules across the membrane in either direction. We saw examples of these in our earlier discussion of alpha helices and beta barrels. We will look at some examples, such as proton pumps and the ATP synthase.
- Enzymes—Many membrane proteins have enzymatic activity for a whole variety of cellular functions. We will explore many examples, including synthesizing or modifying enzymes, flippases, scramblases (shown earlier, in Figure 02-07), or kinases.
- Receptors—Receptors are key for the cell to be able to sense and respond to its environment. Receptors extend across the membrane and bind to small molecules or other proteins on the outside of the cell and in response initiate a chain of events leading to transmission of a signal inside the cell. Many receptors are also enzymes. We will look at receptors in more detail when we discuss cell signaling.
The Plasma Membrane: An Example of a Real Biological Membrane
The plasma membrane is the membrane that surrounds the cell and is the first point of contact between the cell and its environment. Red blood cells were one of the first cells to have their plasma membranes studied and as such have one of the most well-characterized plasma membranes. In most cell types, the plasma membrane is supported and shaped by a network of proteins. In red blood cells, most of the protein network is inside the cell, just underneath the plasma membrane. However, in other cell types, this network can also be on the outside (via connections to the extracellular matrix or cell wall) or on both sides of the membrane.
Figure 02-18 shows a diagram of the plasma membrane of a red blood cell. In it, we see that the lipid bilayer of the plasma membrane is attached to an internal meshwork of spectrin protein filaments. These filaments provide a framework that supports the membrane and gives it its elasticity. You can also see that lipids and proteins on the extracellular side of the membrane are connected to carbohydrates (as indicated by the glyco- prefix). Since red blood cells move through the bloodstream, spectrin does not make any permanent connections with external structures.
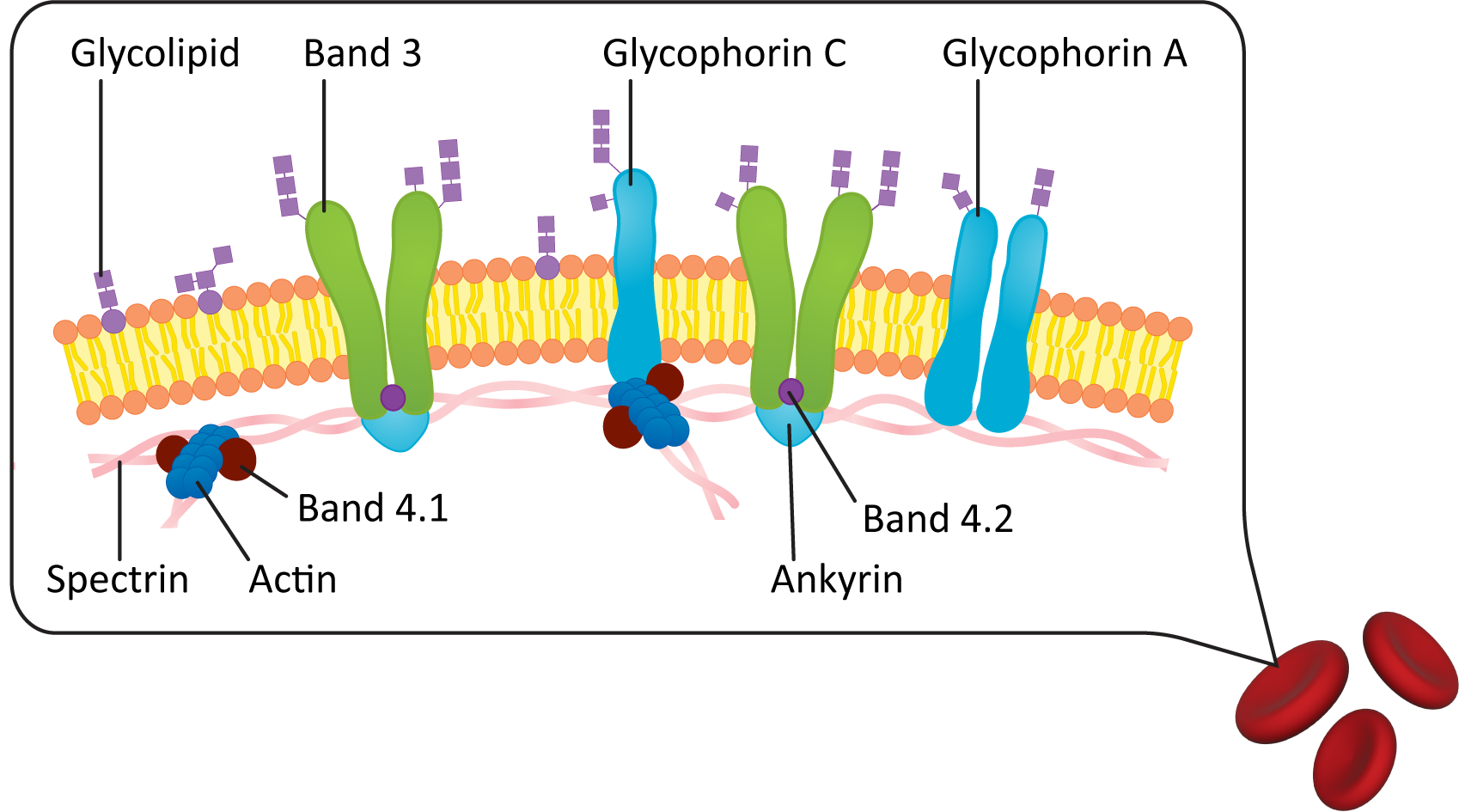
Plasma Membrane Carbohydrate Groups Are Found on the Outside of the Cell
Something that is important to point out in the plasma membrane figures in this chapter (see Figures 02-06, 02-10, and 02-18 as examples) is that the exterior of the plasma membrane is usually covered in a coating of carbohydrates. These carbohydrates are most commonly in the form of glycolipids and glycoproteins that are integrated directly into the plasma membrane. The carbohydrate component of the plasma membrane has a very important function. In biological systems, cells have an identity (and can be recognized) on the basis of the configuration of carbohydrate molecules on the surface of the membrane.
A classic example of this is your blood type. The cell surface polysaccharides carried by your red blood cells are genetically determined so that your body knows which cells belong to you and which are foreign. The ability to identify foreign cells is absolutely vital for your body so that it can recognize pathogens and other invaders. This also influences our ability to carry out blood transfusions when needed. Your “blood type” makes direct reference to the polysaccharides carried on the surface of your cells. The ABO system is the most well known; however, there are other cell surface signals that the body uses to identify which blood belongs to you, such as the Rhesus (Rh) factor. If you receive a transfusion of blood that contains the incorrect polysaccharide markers on the cell surface, your body will produce antibodies to attack the blood and destroy it.
In some cell types (such as bacteria but also many eukaryotic cells), this carbohydrate coating on the plasma membrane is complex enough that it has its own name: the glycocalyx. Most examples of eukaryotic cells that have a glycocalyx are found in animals. This includes many epithelial cells (like the cells lining the gut and our blood vessels). Another really great example of a glycocalyx, which you can actually see with your naked eye, is the slimy coating on fish. The polysaccharide coating is found on virtually all fish, including the sockeye salmon shown in Figure 02-19. It plays multiple roles, including protection, cell-to-cell recognition, and even immune functions.

Plasma Membrane–Adjacent Structures in Animal and Plant Cells
Cells in tissues are surrounded by a matrix of protein, polysaccharide, and fluid. The composition of the extracellular environment varies widely in different tissue types as well as in different organisms from different kingdoms. However, a common theme is that there are quite a lot of carbohydrates. The carbohydrates may or may not be associated with proteins, but collectively they form a three-dimensional network that connects cells together and provides a sort of hydration layer to trap water near the cells. Here we will highlight some of the key differences between this external environment of plant and animal cells.
In both plants and animals, the extracellular macromolecules are synthesized and secreted by the cells that live within them through a process called exocytosis, which we will cover in Chapter 4.
The Extracellular Matrix of Animal Cells
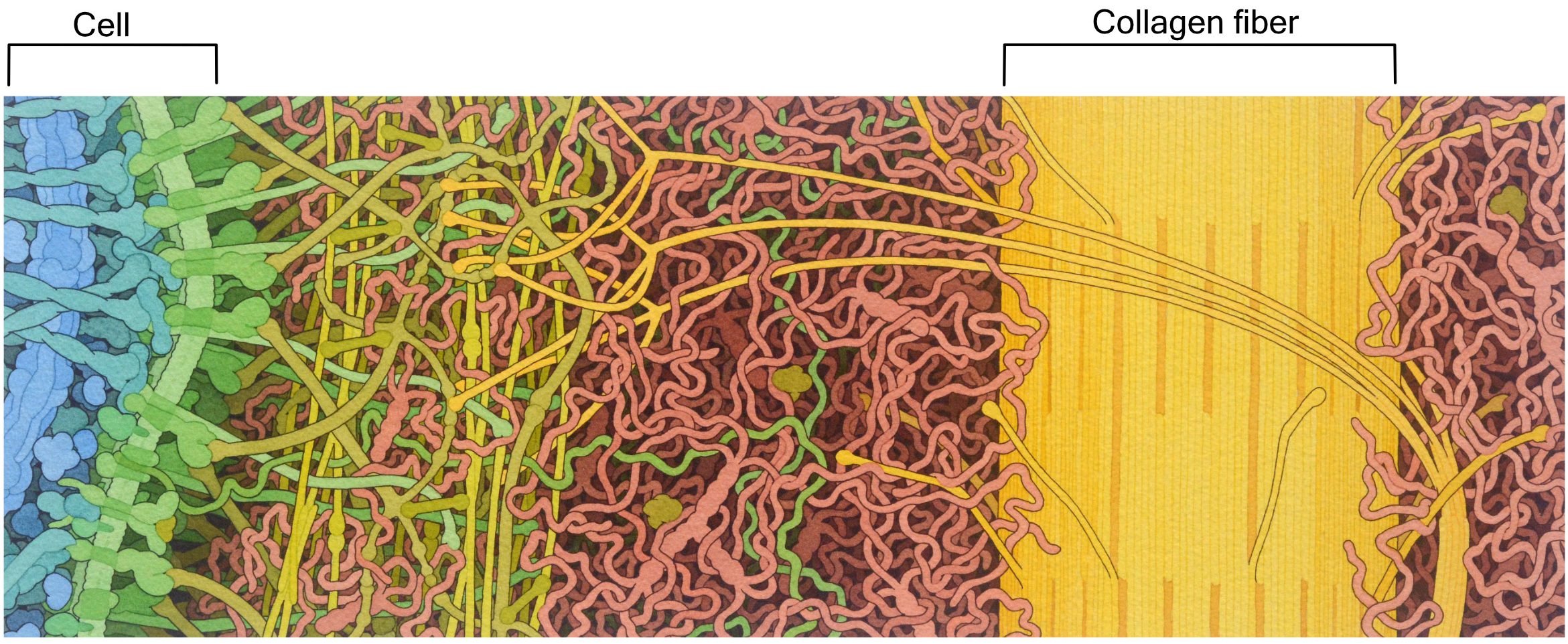
The extracellular matrix (ECM) of animal cells is made primarily of proteoglycans. These are very similar to glycoproteins, except there is quite a lot more polysaccharide attached to a relatively small protein. The proteins and polysaccharides of the extracellular matrix are all connected to each other in a large 3D network, making it difficult to tell where one molecule ends and another begins. Polysaccharide chains known as hyaluronic acid are a major component of the animal extracellular matrix. Hyaluronic acid is important due to its gel-like properties. It helps trap water, which makes the entire ECM look and feel a bit like Jell-O. Thus, polysaccharides like hyaluronic acid help the extracellular matrix remain hydrated and, as such, resist compression. For this reason, it is a large component of the cartilage in our joints.
The most abundant protein in the mammalian extracellular matrix (not to mention the most abundant protein in the entire human body) is a very large protein called collagen (Figure 02-20). This protein is made of three intertwined polypeptide chains, which makes collagen very strong. Collagen provides structural support and helps with things like wound healing.
The Plant Cell Wall
The plant cell wall is also made mostly of carbohydrates. Interestingly, it contains many fewer proteins and is more rigid than an animal extracellular matrix. There is a cell wall that surrounds every single cell in a plant. So while it can be thought of as a type of extracellular matrix for the plant, its role is more complex than that of the animal extracellular matrix. It provides the structural support for the whole plant, similar to the role of the skeleton in animals.
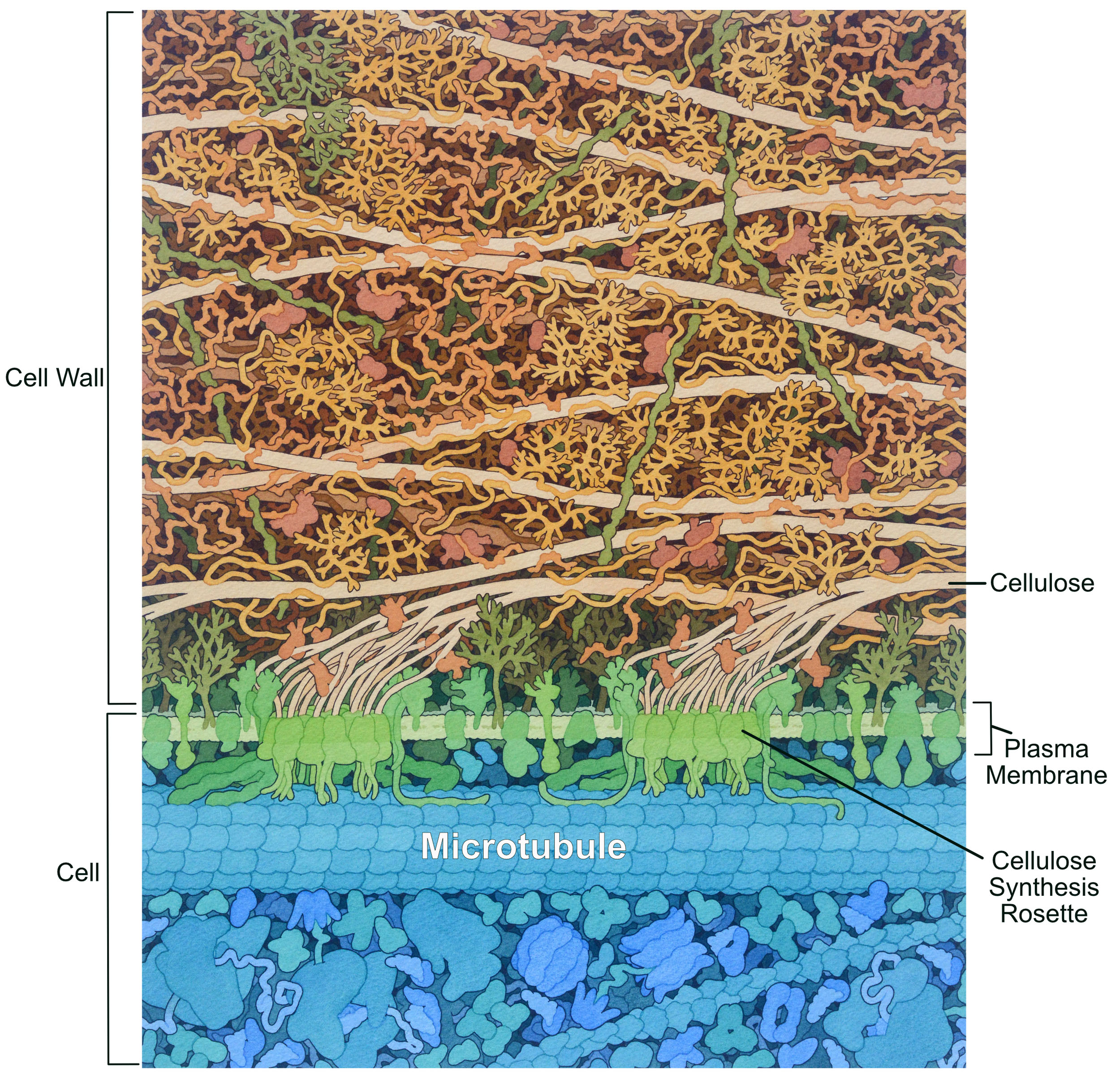
Like the animal extracellular matrix, the plant cell wall is synthesized and secreted by the cell. Most of the polysaccharide components are synthesized by the Golgi apparatus; however, there is one notable exception. Cellulose is a very strong fiber of crystallized glucose chains that is synthesized in a unique structure called a rosette (Figure 02-21). This rosette is embedded in the plasma membrane, and it moves through the membrane as cellulose is synthesized. Microtubules have a role to play in the synthesis of cellulose and, as such, have a very different organization than we see in animal cells.
While there are a few proteoglycans and glycoproteins in the plant cell wall, the complex, branched polysaccharides are really the key players in both its structure and its function. In addition to cellulose, there are other long polysaccharide chains, such as hemicellulose (used to connect the cellulose together) and pectin. Pectin acts in a similar way to hyaluronic acid in that it helps trap water and create a gel matrix. Incidentally, this is also why we use pectin to make jam.
Topic 2.4: Experimental Techniques in Membrane Biology
Learning Goals
- Explore the two most common experimental approaches to studying membrane proteins by analyzing data from representative examples:
- hydropathy plots as an example of the bioinformatics approach to studying proteins
- gel electrophoresis and SDS-PAGE as an example of a biochemical approach to studying proteins
Biological research changed quite drastically in 2000, when the human genome was completely sequenced (see the original publication of the human genome). Since then, our access to genetic information from a variety of species has increased much faster than we could have possibly imagined. This has fundamentally changed how we do research and has revolutionized how we explore cell biology. We now do much initial work in silico (meaning inside the computer) to first make solid predictions about protein function based on recognizable patterns from their primary sequence. To test the predictions made in in silico experiments, we turn to laboratory techniques that manipulate the DNA and proteins directly. In this section, we will explore one bioinformatic technique and one laboratory technique as examples of the many ways that scientists explore biological questions about membranes and their proteins.
Technique 1: Bioinformatics Approaches and the Hydropathy Plot
The amount of DNA sequence information that is openly accessible on the web continues to increase. The data sets can be quite large and can be difficult to handle without the help of computer programs and algorithms to organize, align, and make predictions about structure, function, and subcellular location of unknown proteins. The field of computer-based analysis of DNA and proteins has become so important that it has its own name—we call this bioinformatics.
Protein folding is subject to the same underlying chemistry and thermodynamics as everything else in the universe. This means that as long as we know the primary sequence of a protein, we can make predictions about structure, which can also tell us important information about protein function. These days we use computer algorithms and modeling software to learn as much as we can about a protein before we start to experiment in the lab. In some cases, lab experimentation on a protein is not possible, so computer predictions are our only option. The number of online apps and tools that are available (many for free) also continues to increase as biological computer scientists continue to build tools to answer questions. There are far too many of these tools to reasonably cover in any detail, so we’ll focus on one of the most fundamental: the hydropathy plot.
A hydropathy plot is a bioinformatic tool that analyzes the sequence of amino acids in a protein and looks for specific patterns. Hydropathy plots can go by a few different names in the wider world (hydropathy index, hydrophilicity or hydrophobicity plots, etc.), but they all search for the same thing.
Specifically, hydropathy plots predict alpha-helical transmembrane domains. To do this, they look for a linear stretch of amino acids that are “hydrophobic enough” to allow them to exist stably within the confines of the nonpolar portion of the lipid bilayer. Usually, this equates to around 18–20 nonpolar amino acids in a row in the primary sequence. This is the approximate number of amino acids required to span the length of a typical membrane bilayer when adopting the alpha-helical arrangement. Video 02-04 explains hydropathy plots (called both hydropathy scales and indexes in this video) in more detail.
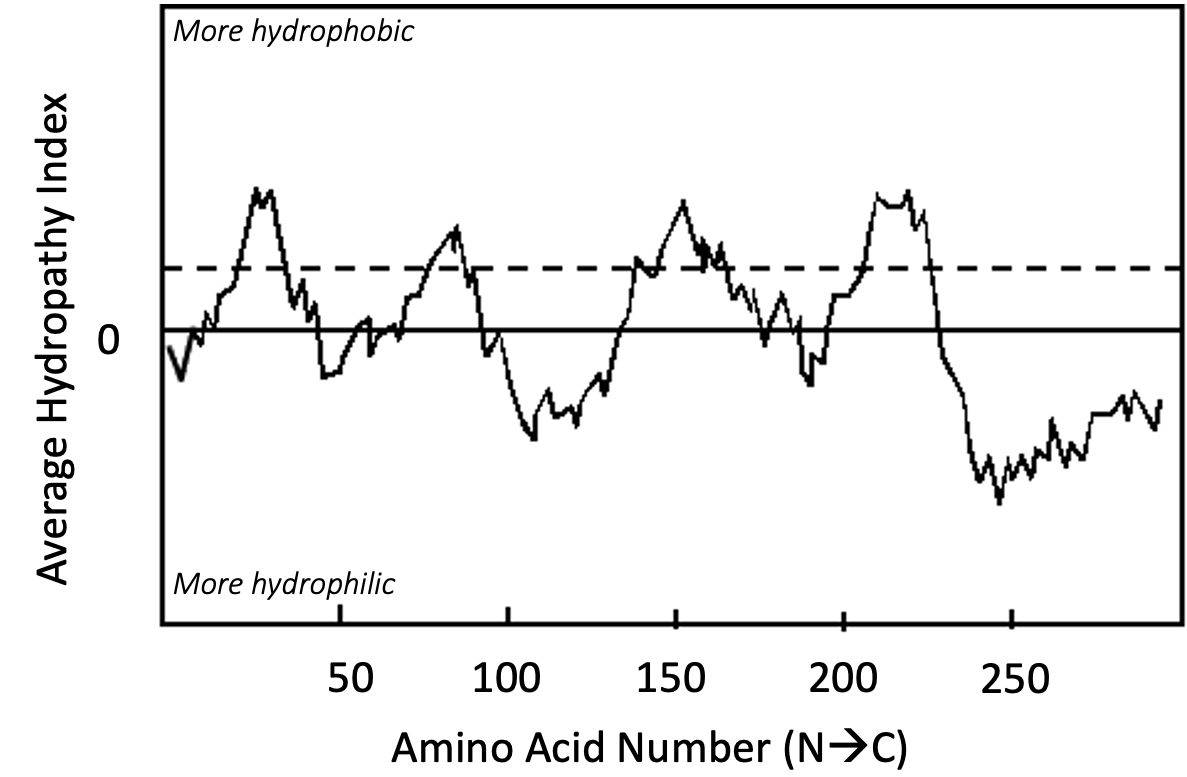
The output of the computer analysis of the primary sequence takes the form of a graph (Figure 02-22), where
- the x-axis is the linear sequence of the protein, listed from N-terminus to C-terminus, and
- the y-axis is a number known as the hydropathy index.
The hydropathy index is a measure of the average hydrophobicity of a given amino acid residue (hydropathy score) and the average hydropathy score of a certain number of adjacent amino acids on either side combined into one value. The hydropathy score of each amino acid is a constant that has been determined based on computer-generated and experimental data. You can think of it as a number that measures how difficult it would be to immerse that amino acid into water. A higher hydropathy score indicates that it takes more energy to immerse that amino acid in water, which indicates it is more nonpolar than other amino acids.
At each point on the graph that the line is above 0, the amino acid and its neighbors are considered to be nonpolar, and below 0, the amino acid and its neighbors are polar on average. If the peak is high enough and wide enough, then a transmembrane alpha helix becomes a reasonable prediction. In this textbook, the “threshold hydrophobicity” is marked on the graph by a dashed line, and anything that crosses that line is considered to be “hydrophobic enough” to be a potential transmembrane region. Note that only major peaks are counted, not minor ones.
It is worth emphasizing that the hydropathy plot can only make predictions about protein structure. In order to confirm the prediction, you would be expected to follow up your bioinformatic analysis with additional experimentation. It is entirely possible that even though this protein is predicted to be transmembrane, the reality could be quite different. The “transmembrane region” predicted here could actually be a region sequestered inside of a soluble protein, or it may have a completely different function entirely. A good scientist will always follow up computer-based predictions with experiments to prove that the computer predictions are true. We will see examples where hydropathy plot predictions are, in fact, false when we discuss the endomembrane system in Chapter 4.
Technique 2: Biochemical Approach: Gel Electrophoresis
Despite the many advantages of bioinformatics approaches, there are also disadvantages that must be considered. For one thing, bioinformatics can only make predictions, and those predictions are limited to what we can learn from the amino acid, DNA, or RNA sequence. At some point, the scientist must test their bioinformatic predictions experimentally. One of the most commonly used techniques in cell and molecular biology is known as gel electrophoresis.
Gel electrophoresis is a way to separate macromolecules based on chemical features like size and charge. We can use gel electrophoresis to separate protein, DNA, or RNA (but not usually all at the same time!). In this technique, molecules are inserted into a gel matrix (not dissimilar from Jell-O but made from a different substance) that has pores of particular size. This gel matrix holds the sample and provides a filter to separate the molecules. The term electrophoresis refers to the electric field that is used to separate the molecules. Since the molecules carry a charge, the electric current causes them to move toward one side of the gel (see Video 02-05 below).
There are many different types of gel electrophoresis techniques available. Each one is named slightly differently to represent its unique features. Here’s a short list of the most common types:
- SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) is used to separate proteins based almost exclusively on their molecular weight. This technique involves dissolving and denaturing proteins with the detergent SDS. This detergent adds a strong negative charge to the protein equal to the number of amino acids. This causes it to migrate to the positive pole during electrophoresis based on the size of the protein.
- Agarose gels can be used to separate nucleic acids. Nucleic acids naturally contain a strong negative charge on each residue, so detergents like SDS are not required. This consistent charge/mass ratio allows these molecules to travel to the positive electrode and separate based on size.
- Native gels do not denature molecules in contrast to SDS-PAGE. Thus, this technique exploits the natural charge carried by proteins, so the size, shape, and natural charge of the folded molecule also impact its movement in the gel.
- 2D gels separate molecules based on molecular weight and isoelectric point.
It’s worth noting that gel electrophoresis on its own doesn’t tell us much. It is merely the tool we use to separate the molecules in our sample. It is important to know how the samples are treated before they are separated on the gel. In addition, in many samples, there are lots of proteins mixed together. Thus, often an additional technique is required afterward SDS-PAGE to highlight and/or identify the proteins of interest in the gel (i.e., staining, antibody labeling, mass spectrometry, etc.). We will first show an example where we can use gel electrophoresis to study proteins using SDS-PAGE. Afterward we will briefly touch on how it can be used for studying nucleic acids in preparation for Chapter 3.
Gel Electrophoresis to Study Proteins: SDS-PAGE
SDS-PAGE is one of the more commonly used types of gel electrophoresis. Its name comes, in part, from the molecules used in the technique. A polyacrylamide gel is a specific type of gel that can be used to separate proteins primarily. SDS is a detergent that helps denature and separate the proteins in your sample. First, a brief video (Video 02-05) shows how the process works.
As you can see, protein samples are first treated with the heat and the detergent SDS (Figure 02-23). The heat helps the proteins to unfold and become linear. The SDS then evenly coats the proteins, which helps them maintain their unfolded state and also adds a uniformly negative charge. Thus, all proteins end up completely unfolded, reducing any variation of travel based on the shape/compactness of their normal folding pattern. This makes it so that proteins are separated by their size and not some other characteristic.
Once the samples are heated and treated with SDS, they are added to the gel matrix using small wells that are set near the negative electrode (i.e., the cathode). The electric current is then turned on, and the proteins travel toward the positive electrode (i.e., the anode) at the other end of the gel. As the current runs through the gel, the proteins move slowly toward the positive electrode. Proteins that are larger will take longer to move through the gel matrix, and smaller proteins will travel farther through the gel. This way, the proteins in the sample are separated over time, with the larger proteins staying closer to the site or origin and small proteins moving farther away.
As mentioned at the start, SDS-PAGE on its own doesn’t tell us much. It is merely a way to separate the proteins in a sample by molecular weight. The experiment you want to do will always include treatments of your samples before you do SDS-PAGE. The animation below (Video 02-06) shows an example of an experiment that uses SDS-PAGE to determine the type of membrane protein found in a sample of plasma membrane.
DNA Gel Electrophoresis
While this chapter is not about nucleic acids and DNA, we wanted to add a brief discussion of how gel electrophoresis can be used to study nucleic acids, for two reasons:
- This is, by far, the most common usage of gel electrophoresis in modern times.
- The next chapter of this textbook looks at DNA and genomes in more detail, and you will need to understand how DNA gels work in order to be prepared for the techniques covered in Chapter 3.
Gel electrophoresis for separating DNA by size follows many of the same principles as SDS-PAGE, but with some differences:
- SDS is not required, as the phosphates in the backbone of DNA create a uniform negative charge on the molecule that is proportional to its size.
- The DNA gets loaded into a gel matrix made of agarose instead of polyacrylamide. DNA and RNA can be quite large—too large for a polyacrylamide matrix. The wider pore sizes in an agarose matrix accommodate the larger molecules.
Like SDS-PAGE, an electric current is run through the gel once the samples are loaded, and the DNA will travel toward the positive terminal. As a result, just like in SDS-PAGE, the DNA will separate based on the size of the molecule such that bigger molecules will stay closer to the origin and smaller particles will move farther away. Video 02-07 is an excellent explanation of gel electrophoresis using agarose gels for DNA. Compare this technique to what you learned about SDS-PAGE above.
As you explore the techniques in Chapter 3, don’t forget to come back here if you need to remember exactly how gel electrophoresis works.
Chapter Summary
In this chapter, we’ve explored the structure and function of membranes. To do this, we first needed to explore some of the characteristics of the lipids and proteins that the membrane is made of. We identified four major characteristics of membranes:
- The membrane is a bilayer made up of lipids and proteins.
- The membrane is selectively permeable.
- The membrane is organized but fluid.
- The membrane is asymmetric.
We also learned that the fluidity of the membrane is dependent on its lipid composition—namely, the length and degree of unsaturation of the phospholipid tails and the cholesterol content. After that we discussed the difference between integral and peripheral proteins and how the secondary structure of membrane proteins contributes to its ability to span the entire membrane, including the hydrophobic portion of the membrane. We rounded out our discussion of membranes by briefly exploring the plasma membrane, which is a unique membrane in the cell, as it is in contact with the external environment. Outside of the plasma membrane is an additional structure known as the extracellular matrix in animal cells and the cell wall in plants and fungi. While these two have some similar roles, they are not the same.
Finally, we explored several experimental techniques throughout this chapter that can help us learn about membranes: fluorescence recovery after photobleaching (FRAP) helps us measure the movement of membrane components; hydropathy plots are a bioinformatic tool that predicts membrane-spanning alpha helices from the primary sequence of a protein, and SDS-PAGE and other kinds of gel electrophoresis can be used to learn more about DNA, RNA, and protein.
Review Questions
Note on usage of these questions: Some of these questions are designed to help you tease out important information within the text. Others are there to help you go beyond the text and begin to practice important skills that are required to be a successful cell biologist. We recommend using them as part of your study routine. We have found them to be especially useful as talking points to work through in group study sessions.
Topic 2.1: The Chemical Features of Biological Membranes
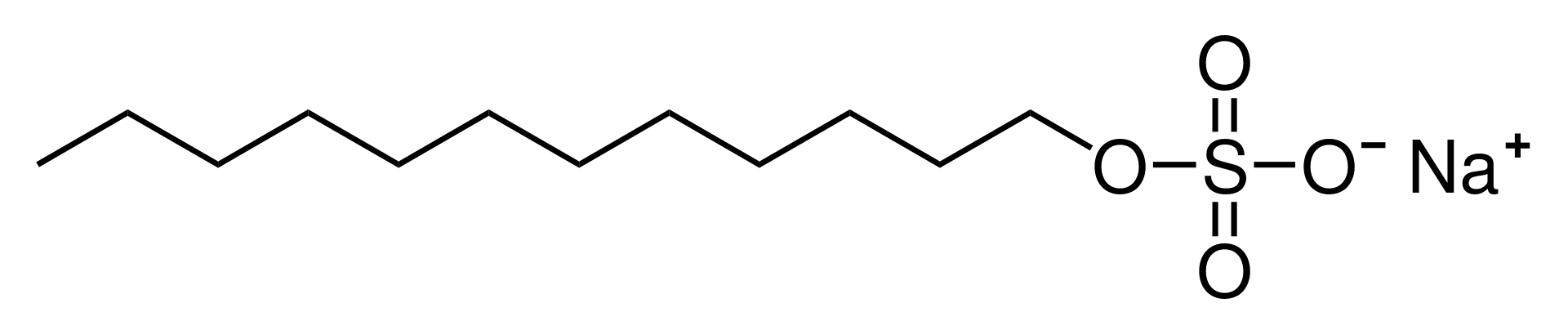
- For the following molecules (phosphatidylcholine, cholesterol, generic glycolipid, sodium dodecyl sulfate [SDS]), examine the molecular structures and label the regions specified below. Note that each region may or may not be present in each molecule:
- the polar region (differentiate between charged and uncharged molecules of the polar region) and the nonpolar region
- a region that would be stiff and inflexible
- a glycerol residue (Some molecules have a serine residue instead; does yours?)
- a region that could easily have C=C bonds added (How would that affect the structure of that region?)
- Identify which of these lipids (phosphatidylcholine, cholesterol, generic glycolipid, sodium dodecyl sulfate [SDS]) would be able to form lipid bilayers on their own. Use the details of the structures that you drew to explain why or why not.
- Explain how thermodynamics, amphipathicity, and shape of lipids allow for the spontaneous assembly of lipids into phospholipid bilayers.
- Which types of molecules are able to cross a membrane bilayer. Can you tell based on the chemical properties of the bilayer compared to the molecules attempting to cross? Explain.
- For each of the four features of the cell membrane, explain how proteins and lipids generally contribute to those features. Where do carbohydrates fit in?
- Explain why it is rare that lipids are able to flip between leaflets in a lipid bilayer.
- What is meant by the term membrane asymmetry and how is this feature created and maintained?
- What are the major differences between a synthetic phospholipid bilayer and a biological membrane?
Topic 2.2: Maintaining Fluidity in the Membrane
- How do cells adjust their membrane composition to maintain fluidity of their lipid bilayers in varying conditions?
- Despite appearances, cholesterol cannot form bilayers on its own. Use the structure of the molecule to explain why.
- Explain how the structure of phospholipids is the basis of the major properties of the bilayers that they form: physical form of the bilayer, self-sealing property, selective permeability, and fluidity of the bilayer.
- Describe how fluorescence recovery after photobleaching (FRAP) works and list the types of scientific questions that can be answered using this technique.
- Describe the composition of lipid rafts and their role in cells.
- Explain how reduced temperature can result in gel phase membranes. Why is this disastrous for living membranes?
- Exposing living membranes to temperatures that are higher than what they are adapted for can also be disastrous… What’s the impact of higher temperatures on membrane fluidity, and why might that be detrimental?
Topic 2.3: Structure and Function of Membrane Proteins
- Find images of all 20 amino acids (don’t memorize them!) online. Critically assess the structures and identify the following:
- The portion of the amino acid that is common to all of them.
- The portion that is unique to each amino acid, known as the R group or side chain.
- The functional groups that will form the peptide bond. Will anything be lost/gained during that reaction? How do these parts relate to the “N” and “C” terminus of a protein?
- The next questions are specifically for the R groups. Based solely on their structure, identify the following:
- Any R groups that you would expect to have acidic properties. Will they gain or lose a proton during that reaction?
- Any R groups that you would expect to have basic properties. Will they gain or lose a proton during that reaction?
- Any R groups that would not be able to form H-bonds with water.
- R groups that would form H-bonds but would not be acidic or basic.
- R groups that could interact ionically with their neighbors.
- Any R groups you would consider to be “big” or “small” relative to the others.
- Define the four levels of protein folding and explain how each one is stabilized.
- Are disulfide bridges covalent or noncovalent interactions? How do they form? Why are they considered to be uncommon in the cytosol?
- Describe the functional roles of the following examples of membrane proteins:
- Integral proteins
- Peripheral proteins
- Glycoproteins
- What is the difference between integral and peripheral membrane proteins? Discuss and compare the different strategies for association of proteins with membranes.
- What is a domain in a protein? How does it relate to structure and/or function?
- Compare the hydrophobic forces that hold a membrane protein in the lipid bilayer to those that help proteins fold into a unique three-dimensional structure.
- How do cells restrict the movement of membrane proteins? Outline the different strategies and provide brief examples.
- What feature of a membrane’s structure allows it to have separate identities and functions on each side?
Topic 2.4: Experimental Techniques in Membrane Biology
- What is a hydropathy plot, and how does it help predict transmembrane regions?
- Explain why hydropathy plots can only make predictions about transmembrane alpha helices and not beta barrels.
- Explain why experimental techniques are needed to verify predictions from hydropathy plots.
- Why is SDS needed for sample preparation in SDS-PAGE experiments?
- What types of questions can be answered using SDS-PAGE?
- How is DNA gel electrophoresis different from SDS-PAGE on proteins?
A selectively permeable barrier that is designed to separate the cell from the external environment, allows communication of activities between cells, and functions to form intracellular compartments. It is composed of lipids, proteins, and carbohydrates.
Composed of two sheets of phospholipids. The lipids are orientated so that the polar heads are facing outward and the nonpolar fatty acid tails are facing inward.
Refers to molecules that have one end of the molecule that is different from the other end. Most often, this refers to an uneven electron distribution, where extreme uneven distribution of electrons results in ionized molecules with a charge. Polar can also refer to shape differences on different ends of a molecule, as is the case with polar cytoskeletal elements.
Refers to molecules that have do not have one end of the molecule that is different from the other end. Most often, this refers to electron distribution. Nonpolar molecules share electrons evenly and never carry a dipole or charge. Nonpolar can also refer to the symmetrical shape of molecules, such as the tetrameric form of intermediate filaments.
“A class of lipids whose molecule has a hydrophilic ‘head’ containing a phosphate group and two hydrophobic ‘tails’ derived from fatty acids, joined by an alcohol residue (usually a glycerol molecule).” Source: Phospholipid. (2023, May 25). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Phospholipid
Generally refers to a lipid bilayer that has two lipids stacked tail to tail with the polar head groups facing the aqueous environment.
A chemical compound or molecule that possesses both a hydrophilic region and a hydrophobic region. Phospholipids that make up the majority of the structure of phospholipid bilayers are an example of an amphipathic molecule.
Latin for “water loving.” Refers to chemical groups that readily associate with polar molecules like water.
Latin for “water fearing.” Refers to chemical groups that do not readily associate with polar molecules like water. Instead, these are generally nonpolar in nature and group with other nonpolar molecules.
A single layer of a lipid bilayer.
“A small artificial vesicle, spherical in shape, having at least one lipid bilayer.” Source: Intermolecular force. (2023, June 22). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Intermolecular_force
“The observed tendency of nonpolar substances to aggregate in an aqueous solution and exclude water molecules.” Source: Hydrophobic effect. (2023, June 13). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Hydrophobic_effect
The Gibbs free energy equation. When the ΔG is negative, this indicates a spontaneous reaction and does not need extra energy input to occur.
A feature of biological membranes that allows only certain molecules and ions to pass through to the inside of the cell. Selective permeability is useful in controlling the composition of the internal cellular environment.
Refers to proteins in a membrane that facilitate the movement of molecules across a biological membrane.
A family of lipids containing a phosphate head, two fatty acid tails, and an inositol head group. These compose a small but functionally relevant component of the cytosolic side of a cell’s membranes.
“A glycoprotein and glycolipid covering that surrounds the cell membranes of bacteria, epithelial cells, and other cells.” Source: Glycocalyx. (2023, July 9). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Glycocalyx
“A structural layer surrounding some types of cells, just outside the cell membrane. It can be tough, flexible, and sometimes rigid. It provides the cell with both structural support and protection, and also acts as a filtering mechanism.” Source: Cell wall. (2023, March 30). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Cell_wall
“A network consisting of extracellular macromolecules and minerals, such as collagen, enzymes, glycoproteins and hydroxyapatite that provide structural and biochemical support to surrounding cells.” Source: Extracellular matrix. (2023, June 2). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Extracellular_matrix
A protein that will move lipids from one monolayer (leaflet) to the other in a lipid bilayer. They are not selective in direction and do not require an energy input.
“Transmembrane lipid transporter proteins located in the membrane that belong to ABC transporter or P4-type ATPase families. They are responsible for aiding the movement of phospholipid molecules between the two leaflets that compose a cell’s membrane (transverse diffusion, also known as a ‘flip-flop’ transition).” Source: Flippase. (n.d.). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Flippase
A method to determine the mobility of molecules within a membrane. A region of the cell is photobleached (the fluorescent molecules are destroyed). The same area is monitored over time to determine if fluorescence returns to the bleached area.
A lipid that has at least one double bonded carbon in its chain.
A lipid that has no double bonds within the hydrocarbon chain.
A molecule in the sterol family that plays a role in the fluidity of membranes.
A distinct membrane region that contains specialized lipids and proteins. Often these regions are a bit thicker than surrounding membrane areas and provide specialized functions like cell signaling.
Refers to fading. Often this is in reference to a fluorescence molecule that is no longer able to be seen.
The graph formed by a FRAP experiment. It measures the fluorescence intensity of a region over time to assess how much fluorescence “recovers” after the initial photobleaching step.
A variable chemical structure off the central carbon of an amino acid. The different chemical features of the R group define each amino acid and give it its overall characteristics (size, shape, charge etc.).
The repeating atomic structure formed when amino acids are joined through a peptide bond. These form N-C-C repeats referring to the central atoms in the amide, R-group, carboxylic acid groups that form each amino acid.
Refers to the sequence of the protein. Amino acids are joined into a polymer. The unique sequence of each polypeptide chain dictates how it will fold.
“The force that mediates interaction between molecules, including the electromagnetic forces of attraction or repulsion which act between atoms and other types of neighboring particles, e.g., atoms or ions. Intermolecular forces are weak relative to intramolecular forces—the forces which hold a molecule together.” Source: Intermolecular force. (2023, June 22). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Intermolecular_force
“Proteins that assist the conformational folding or unfolding of large proteins or macromolecular protein complexes.” Source: Chaperone (protein). (2023, April 2). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Chaperone_(protein)
Refers to local repeated backbone-backbone interactions of the polypeptide chain. There are two common forms of secondary structure: alpha helices and beta sheets.
“A common motif in the secondary structure of proteins and is a right hand helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence.” Source: Alpha helix. (2023, July 19). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Alpha_helix
“A common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation.” Source: Beta sheet. (2022, September 9). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Beta_sheet
All the structure of the protein that is not primary or secondary or quaternary. This defines the 3D shape and encompasses interactions between side chains, side chains–backbone, and backbone-backbone (that are not local or repeated).
Refers to the interactions between two independent polypeptide chains. Not all proteins have quaternary structure. Some will only have quaternary structure transiently.
The covalent linkage of thiol groups between two thiol residues. Commonly, these are found on the R groups of cysteine amino acids.
A type of protein that is embedded into biological membranes. Transmembrane proteins are an example of integral membrane proteins that span the entire biological membrane and are used to transport material from one side to the other.
Membrane proteins that do not embed in the biological membrane. Instead, they bind to other integral proteins or more rarely interact with the lipids themselves.
A protein that has a region that extends through the full membrane. As a result, the protein will have exposure to both sides of the membrane.
A protein that is embedded in a biological membrane but only interacts with one layer of the bilayer.
“A beta sheet composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand (hydrogen bond). Beta-strands in many beta barrels are arranged in an antiparallel fashion. Beta barrel structures are named for resemblance to the barrels used to contain liquids.” Source: Beta barrel. (2022, May 5). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Beta_barrel
A biological membrane that separates the cell from the surrounding environment. It is composed of a phospholipid bilayer, proteins, and carbohydrate components.
“Proteins that are heavily glycosylated. The basic proteoglycan unit consists of a ‘core protein’ with one or more covalently attached glycosaminoglycan (GAG) chain(s).” Source: Proteoglycan. (2023, February 2). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Proteoglycan
A carbohydrate polymer often found in the extracellular matrix and often serves a lubrication function.
“The main structural protein in the extracellular matrix found in the body’s various connective tissues.” Source: Collagen. (2023, July 3). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Collagen
A special protein structure where cellulose is made in plant cells destined for use in the cell wall.
Refers to experiments conducted in a computer; computer simulations.
“An interdisciplinary field of science that develops methods and software tools for understanding biological data, especially when the data sets are large and complex.” Source: Bioinformatics. (2023, July 7). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Bioinformatics
A graph used to predict the number of position of transmembrane domains in an amino acid sequence. Each amino acid’s hydropathy score is plotted with the N terminus of the protein starting at position 0 on the x axis. A peak on this plot above the threshold indicates a potential transmembrane domain.
An index (reported on the y axis) that is a measure of the average hydrophobicity of a given amino acid residue (hydropathy score) and the average hydropathy score of a range of amino acids on either side combined into one value.
A set value indicating the hydrophobicity of a given amino acid.
“A method for separation and analysis of biomacromolecules (DNA, RNA, proteins, etc.) and their fragments, based on their size and charge.” Source: Gel electrophoresis. (2023, July 17). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Gel_electrophoresis
A method to separate molecules (primarily proteins but sometimes DNA) by size. For protein separation, SDS (a detergent molecule) helps denature (unfold) the protein and gives it a consistent size-to-charge ratio. Then the proteins are loaded into a gel matrix and induced with an electric charge. The negatively charged proteins move toward the positive terminal. Larger molecules take longer to move through the gel and stay at the top, while smaller molecules can move more easily through the gel and reside at the bottom.
A method of gel electrophoresis used in biochemistry, molecular biology, genetics, and clinical chemistry to separate a mixed population of macromolecules such as DNA or proteins in a matrix of agarose, one of the two main components of agar. Source: Gel electrophoresis. (2023, July 17). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Gel_electrophoresis
“Native gels are run in non-denaturing conditions so that the analyte’s natural structure is maintained. This allows the physical size of the folded or assembled complex to affect the mobility, allowing for analysis of all four levels of the biomolecular structure.” Source: Gel electrophoresis. (2023, July 17). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Gel_electrophoresis
“A form of gel electrophoresis commonly used to analyze proteins. Mixtures of proteins are separated by two properties in two dimensions on 2D gels.” Source: Two-dimensional gel electrophoresis. (2023, March 2). In Wikipedia, The Free Encyclopedia. https://en.wikipedia.org/wiki/Two-dimensional_gel_electrophoresis

