1 Background
Proteins play vital roles in most biological processes; these roles include acting as catalysts for physiological reactions, as regulators for those reactions, or as structural framework around which these processes can occur. Proteins’ complex organization of diverse functionality in 3D space leads to an astonishing range of function for living organisms. Understanding this intimate relationship between structure and function is the backbone of understanding the natural world and is the key to controlling it.
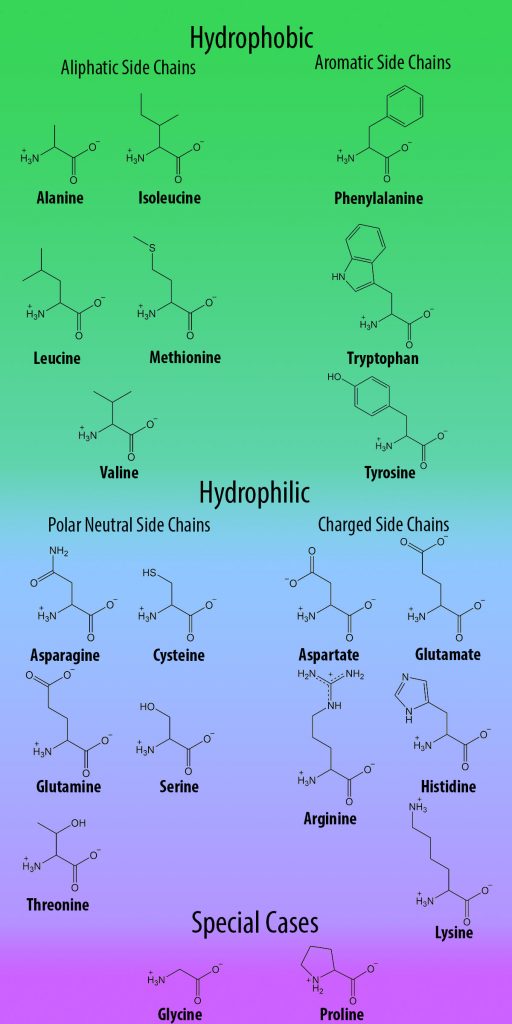
Site-directed mutagenesis (replacing one amino acid codon in a gene with another one) is a common and useful tool for investigating protein function. However, despite their functional complexity, proteins have a relatively simple structural basis: all organisms use the same 20 amino acids to build proteins, all of which have limited chemical functionality (Figure 1). Therefore, in order to expand the range of studies that can be performed on proteins, non-canonical amino acids (ncAAs; also referred to as unnatural amino acids, UAAs) with unlimited chemical functionality are obvious tools for studying these structure-function relationships.
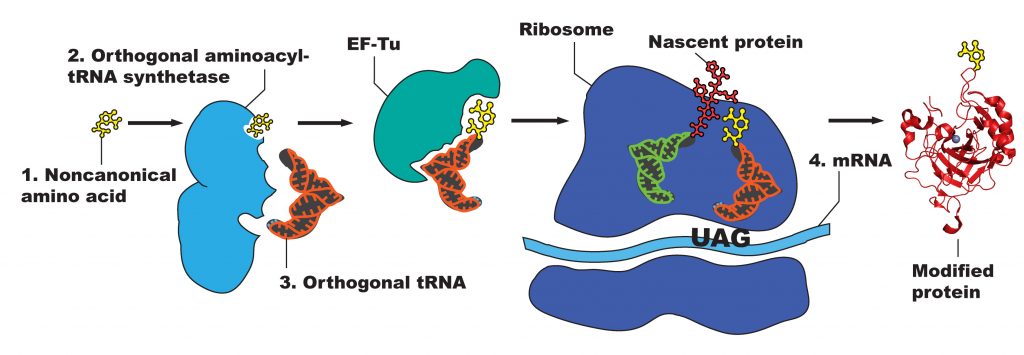
Manipulating proteins to permit use of ncAAs involves exploiting the central dogma of molecular biology —the transcription of a DNA sequence into mRNA, and the translation of this mRNA sequence into a foldable, functional peptide chain. Modification at the DNA level, therefore, will result in a modified protein. The nature of the genetic code, however—with multiple degenerate codons specifying the same amino acid—leaves no “unused” codons for easily adding ncAAs in vivo. Therefore, a codon must be “hijacked” so that it can be used to specify the ncAA by replacing a natural codon in a sequence. The stop codon TAG, the least used of the three stop codons, serves this role and can easily replace any existing codon along a sequence via site-directed mutagenesis. In order to preclude this codon from signifying the termination of the peptide chain, it must function as other the “natural” codons do: having an aminoacyl-tRNA synthetase (RS) bind an amino acid to a custom tRNA, which then adds the amino acid to the peptide chain via the ribosome (Figure 2). Since the amino acid is not naturally occurring, no corresponding RS/tRNA pair is naturally present in the cell.
To facilitate specific ncAA incorporation, the cell must therefore be provided with a ncAA-RS that aminoacylates the paired tRNA with only the ncAA, but no endogenous amino acids. To do this, first a RS and a tRNA must be found in nature that can be used by (are orthogonal to) a host cell. This pair must be able to work both together and with the ribosome of E. coli, and the tRNA must be able to site-specifically recognize the TAG codon. A new, non-toxic ncAA can be chosen for the interesting chemical properties that it may introduce into the protein. Then an RS must be selected that is able to bind the ncAA of interest when aminoacylating the tRNA. Directed evolution of RSs involves rounds of positive and negative selection on a library of RSs (up to 108 members), which all share the same overall structure but have a variety of mutations focused around the amino acid binding site. Positive selection rounds involve transforming library members into cells containing a plasmid with the chloramphenicol acetyltransferase gene (CAT), which confers chloramphenicol resistance, containing a TAG codon. The cells are then grown in the presence of both the ncAA and the antibiotic chloramphenicol; the members that can successfully incorporate the ncAA (as well as those that incorporate endogenous amino acids) produce fully functional CAT protein and can survive in the chloramphenicol-containing medium. To exclude the remaining members that used an endogenous amino acid instead of the ncAA, a round of negative selection is then performed. Negative selection rounds involve isolating the RS-containing plasmids from surviving positive selection cells, then transforming the remaining RS members into cells with another plasmid. This plasmid contains the toxic barnase gene with a TAG codon in the middle, which encodes a toxic protein that kills the cell if successfully produced. ncAA is excluded from the media in negative selection rounds, so that RS members that incorporate a natural amino acid in response to the TAG codon do not survive. Several alternating rounds of positive and negative selection are performed until the remaining RSs are those that can efficiently attach a ncAA to a tRNA (and therefore make protein containing a TAG codon), but cannot attach a natural amino acid to that tRNA.
In order for the cell to produce mutant TAG protein and its necessary ncAA-RS/tRNA pair, it must be provided with the genes that encode them. The genes are introduced into E. coli cells via the plasmids pBad and pDule (Figure 3).
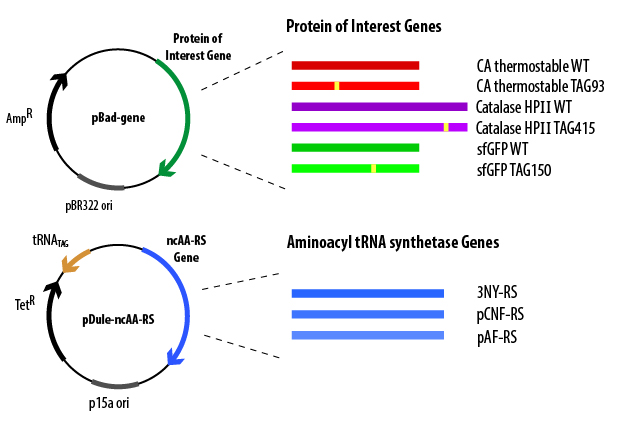
The pBad plasmid contains the gene (containing a TAG codon) that encodes the protein of interest controlled by an arabinose-induced promoter, an origin of replication, and a gene that encodes Β-lactamase (which confers ampicillin resistance). When cloned into the pBad plasmid, codons for 6 histidines are added to either the N- or C-terminus of the gene, which allows for affinity purification of the overexpressed protein. The pDule plasmid contains genes that encode the ncAA-RS and the tRNACUA, an origin of replication (must be compatible with the pBAD origin of replication), and a gene that encodes TetA protein (which confers tetracycline resistance). These plasmids must both be transformed into the cell in order for full-length, ncAA-containing protein to be produced. To verify that the cells contain both plasmids before initiating overexpression, the cells are grown in the presence of the antibiotics ampicillin and tetracycline, ensuring that only cells that contain both plasmids will be able to grow in the media.
Cells containing the appropriate plasmids can then be induced to overexpress the protein of interest using arabinose autoinduction media. The pBad plasmid contains an arabinose promoter system that activates expression of the gene on that plasmid in the presence of arabinose. The autoinduction media is designed to allow cells to reach high density before overexpression is induced, so that a larger number of cells are available to overexpress protein when induction begins. The media autoinduces by using defined sugar concentrations: when the glucose levels begin to decrease due to cellular metabolism and growth, the cells begin to uptake the arabinose that is available. Although they cannot metabolize it for further growth, the arabinose functions as an activator for the promoter on the pBad plasmid, thereby inducing protein overexpression. Crude protein gel electrophoresis can then easily verify the success of this process by monitoring the size of protein (full-length vs. truncated) produced in the presence and absence of ncAA. Full-length protein produced in the presence of ncAA, and truncated protein produced in the absence of ncAA, would indicate that the ncAA-RS/tRNACUA system was able to recognize the TAG codon and incorporate the ncAA, but not any endogenous amino acids.
The quality of a ncAA-protein study is defined by what can be understood about the protein’s structure or function, or by the new ability conferred due to the ncAA’s presence in the protein. Because ncAAs expand the limited chemical potential of amino acid residues, a wide variety of studies and applications become feasible that explore the sensitive yet resilient nature of proteins when using ncAAs. Depending upon the location and type of amino acid substitution, a wide range of effects on protein stability and/or function may occur: both may be unaffected, the stability may be unaffected but activity destroyed, or the protein may not even fold properly, thereby destroying function. Some ncAAs placed in the enzyme active site have even been shown to improve the function of the enzyme by altering the electrostatics of binding or catalysis. If the stability and function of the protein appear largely unperturbed by the addition of the ncAA, then the chemical properties of the ncAA may be utilized to yield relevant information about the structural states of the protein. Examples of different ncAAs and their applicable studies are outlined in Table 1.
Table I. Some ncAA families and their applications
Whether the function of the protein is improved or better elucidated, or the structure and dynamics of the enzyme are better understood, the use of ncAAs has enabled a greater range of studies on protein structure-function relationships.
| Category of ncAA | Example Structure | Applications |
| Photocrosslinking | 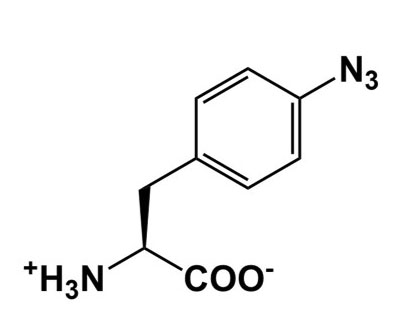 |
Provide snapshots of in vivo protein interactions |
| Biorthogonal ligation |  |
Conjugate fluorophores; surface functionalization |
| Size and polarity probes | 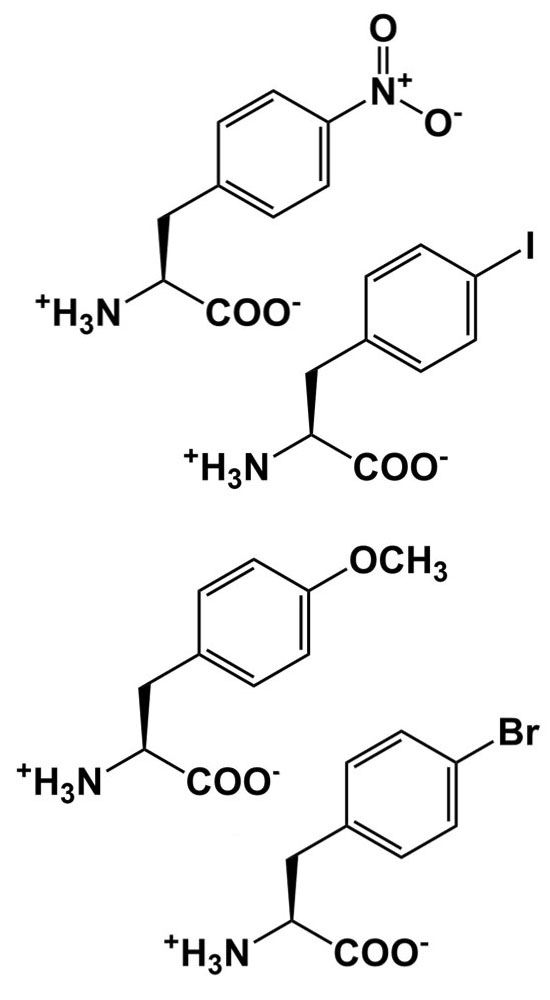 |
Alter packing, sterics, and other interactions to probe structure/function relationships |
| pH probe | 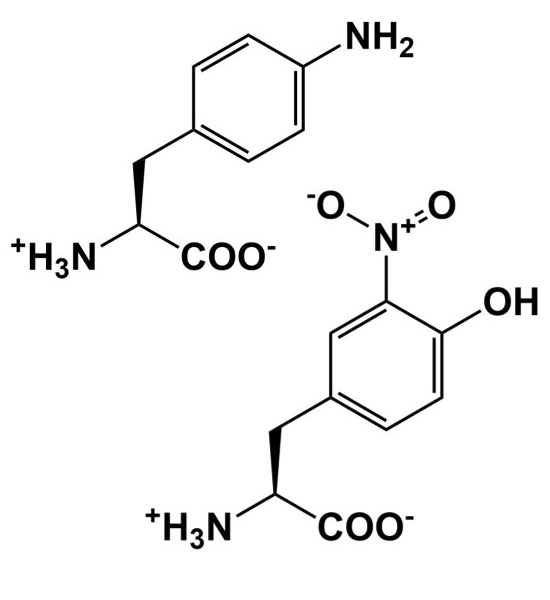 |
Add, remove, and alter hydrogen bonding interactions to study structure/function relationships |
|
Deep Thoughts on Deep Thoughts Throughout this manual, you will find asides, entitled “Deep Thoughts,” on a variety of different topics. These are intended to guide you to develop a deeper understanding of your project and of the techniques you are using as well as to provide some things to consider when designing your experiment, analyzing your results, and carrying out various steps of this laboratory. These suggestions are by no means exhaustive but are intended to help you think critically about what you are doing and guide your design and thought process. We expect that you have read, considered, and answered these questions on your own before asking the instructor or TA for help. |

